

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122160.5 - phase: 0
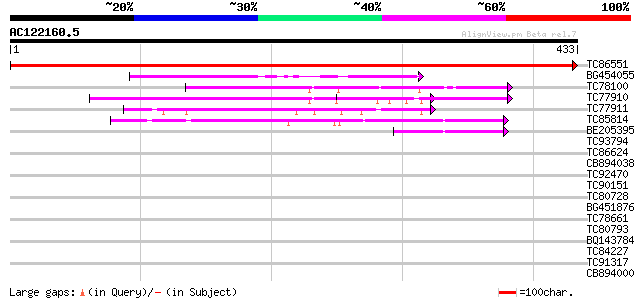
(433 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86551 similar to SP|P48628|FD6C_SOYBN Omega-6 fatty acid desat... 922 0.0
BG454055 similar to GP|5821436|dbj| chloroplast w6 desaturase {C... 131 6e-31
TC78100 similar to SP|P32291|FD3E_PHAAU Omega-3 fatty acid desat... 128 5e-30
TC77910 similar to GP|23345021|gb|AAN17502.1 omega-3 fatty acid ... 102 3e-22
TC77911 similar to GP|23345021|gb|AAN17502.1 omega-3 fatty acid ... 100 8e-22
TC85814 similar to SP|P48631|FD62_SOYBN Omega-6 fatty acid desat... 91 7e-19
BE205395 homologue to SP|P48631|FD62 Omega-6 fatty acid desatura... 42 6e-04
TC93794 homologue to GP|17064784|gb|AAL32546.1 temperature-sensi... 36 0.025
TC86624 similar to PIR|A84900|A84900 hypothetical protein At2g46... 36 0.025
CB894038 31 1.1
TC92470 similar to GP|17065290|gb|AAL32799.1 Unknown protein {Ar... 30 1.8
TC90151 similar to GP|16323326|gb|AAL15376.1 At2g02370/T16F16.16... 29 3.1
TC80728 similar to GP|15810469|gb|AAL07122.1 unknown protein {Ar... 29 4.0
BG451876 weakly similar to GP|15144511|gb| unknown {Lycopersicon... 28 5.3
TC78661 similar to PIR|T09886|T09886 hypothetical protein T22A6.... 28 5.3
TC80793 similar to SP|Q9LUA9|COLE_ARATH Zinc finger protein cons... 28 6.9
BQ143784 weakly similar to SP|P33485|VNUA Probable nuclear antig... 28 6.9
TC84227 weakly similar to GP|6682247|gb|AAF23299.1| unknown prot... 28 6.9
TC91317 similar to PIR|C96761|C96761 hypothetical protein T9L24.... 28 9.0
CB894000 similar to GP|7108597|gb| K+ transporter HAK5 {Arabidop... 28 9.0
>TC86551 similar to SP|P48628|FD6C_SOYBN Omega-6 fatty acid desaturase
chloroplast precursor (EC 1.14.99.-). [Soybean] {Glycine
max}, partial (84%)
Length = 1629
Score = 922 bits (2382), Expect = 0.0
Identities = 433/433 (100%), Positives = 433/433 (100%)
Frame = +3
Query: 1 GSSFQRPILNLRTNISAPYSPGLFNLKGDGLIHKGFRHQSQKHLTPRNKVTVIRAVAIPV 60
GSSFQRPILNLRTNISAPYSPGLFNLKGDGLIHKGFRHQSQKHLTPRNKVTVIRAVAIPV
Sbjct: 69 GSSFQRPILNLRTNISAPYSPGLFNLKGDGLIHKGFRHQSQKHLTPRNKVTVIRAVAIPV 248
Query: 61 EPAPVESAEYRKQLAERYGFEQIGEPLPDNVTLKDVITSLPKKVFEIDDVKAWKSVLISA 120
EPAPVESAEYRKQLAERYGFEQIGEPLPDNVTLKDVITSLPKKVFEIDDVKAWKSVLISA
Sbjct: 249 EPAPVESAEYRKQLAERYGFEQIGEPLPDNVTLKDVITSLPKKVFEIDDVKAWKSVLISA 428
Query: 121 TSYALGLLMISKAPWYLLPLAWAWTGTAVTGFFVIGHDCAHKSFSTNKLVEDIVGTLAFL 180
TSYALGLLMISKAPWYLLPLAWAWTGTAVTGFFVIGHDCAHKSFSTNKLVEDIVGTLAFL
Sbjct: 429 TSYALGLLMISKAPWYLLPLAWAWTGTAVTGFFVIGHDCAHKSFSTNKLVEDIVGTLAFL 608
Query: 181 PLIYPYEPWRFKHDKHHAKTNMLKEDTAWQPVWQDEFESSPFLRKAIIYGYGPFRCWMSI 240
PLIYPYEPWRFKHDKHHAKTNMLKEDTAWQPVWQDEFESSPFLRKAIIYGYGPFRCWMSI
Sbjct: 609 PLIYPYEPWRFKHDKHHAKTNMLKEDTAWQPVWQDEFESSPFLRKAIIYGYGPFRCWMSI 788
Query: 241 AHWLVSHFDLKTFRPNEIKRVKISLACVFAFIAIGWPLIIYKTGIMGWIKFWLMPWLGYH 300
AHWLVSHFDLKTFRPNEIKRVKISLACVFAFIAIGWPLIIYKTGIMGWIKFWLMPWLGYH
Sbjct: 789 AHWLVSHFDLKTFRPNEIKRVKISLACVFAFIAIGWPLIIYKTGIMGWIKFWLMPWLGYH 968
Query: 301 FWMSTFTMVHHTAPHIPFKNSEEWNAAQAQLNGTVHCAYPQWIEILCHDINVHIPHHISP 360
FWMSTFTMVHHTAPHIPFKNSEEWNAAQAQLNGTVHCAYPQWIEILCHDINVHIPHHISP
Sbjct: 969 FWMSTFTMVHHTAPHIPFKNSEEWNAAQAQLNGTVHCAYPQWIEILCHDINVHIPHHISP 1148
Query: 361 KIPSYNLRAAHKSIQENWGKYLNEASWNWRLMKTIMTVCHVYDKQQNYVSFDEVAPKESR 420
KIPSYNLRAAHKSIQENWGKYLNEASWNWRLMKTIMTVCHVYDKQQNYVSFDEVAPKESR
Sbjct: 1149KIPSYNLRAAHKSIQENWGKYLNEASWNWRLMKTIMTVCHVYDKQQNYVSFDEVAPKESR 1328
Query: 421 PIRFLKKVMPDYA 433
PIRFLKKVMPDYA
Sbjct: 1329PIRFLKKVMPDYA 1367
>BG454055 similar to GP|5821436|dbj| chloroplast w6 desaturase {Chlamydomonas
sp. W80}, partial (5%)
Length = 672
Score = 131 bits (329), Expect = 6e-31
Identities = 70/225 (31%), Positives = 119/225 (52%)
Frame = +3
Query: 92 TLKDVITSLPKKVFEIDDVKAWKSVLISATSYALGLLMISKAPWYLLPLAWAWTGTAVTG 151
++ ++ LPK+VF+ KA S++ + T LG+++++ PWYLLP+ W + GTA+TG
Sbjct: 18 SMNELADLLPKEVFQTSTKKAIFSLVKTITFVTLGVVILAYTPWYLLPIGWMFLGTAITG 197
Query: 152 FFVIGHDCAHKSFSTNKLVEDIVGTLAFLPLIYPYEPWRFKHDKHHAKTNMLKEDTAWQP 211
+ + C+ +F NK + DI+GTL LPL+ P++ W+ K++ + +L P
Sbjct: 198 LLTVSYACSKGAFVENKALNDIIGTLCMLPLMTPFQSWQ----KYYNNSRLLS-----SP 350
Query: 212 VWQDEFESSPFLRKAIIYGYGPFRCWMSIAHWLVSHFDLKTFRPNEIKRVKISLACVFAF 271
W +SP W I + + +L+ KR +++ ++AF
Sbjct: 351 FW---VVASP---------------WQWIKAQVAPYINLE-------KRAMVNMILIYAF 455
Query: 272 IAIGWPLIIYKTGIMGWIKFWLMPWLGYHFWMSTFTMVHHTAPHI 316
+ I +PL+ G G +K++LMPWL YHFW S+F H PH+
Sbjct: 456 VCIFFPLMALNLGWWGLVKYYLMPWLVYHFWASSFQKAHQ-LPHV 587
>TC78100 similar to SP|P32291|FD3E_PHAAU Omega-3 fatty acid desaturase
endoplasmic reticulum (EC 1.14.99.-), partial (96%)
Length = 1343
Score = 128 bits (321), Expect = 5e-30
Identities = 79/261 (30%), Positives = 124/261 (47%), Gaps = 11/261 (4%)
Frame = +2
Query: 135 WYLLPLAWAWTGTAVTGFFVIGHDCAHKSFSTNKLVEDIVGTLAFLPLIYPYEPWRFKHD 194
W P+ W GT FV+GHDC H SFS + L+ +VG ++ PY WR H
Sbjct: 326 WIFWPIYWISQGTMFWALFVLGHDCGHGSFSNSSLLNSLVGHFLHSSILVPYHGWRISHR 505
Query: 195 KHHAKTNMLKEDTAWQPVWQDEFESSPFLRKAI-------IYGYGPFRCWMSIAHWLVSH 247
HH +++D +W P+ + ++S + K + I+ Y PF W SH
Sbjct: 506 THHQNHGHVEKDESWVPLTEKMYKSLDNMTKTMRFTFPFPIFAY-PFYLWNRSPGKEGSH 682
Query: 248 FD--LKTFRPNEIKRVKISLACVFAFIAIGWPLIIYKTGIMGWIKFWLMPWLGYHFWMST 305
F+ K F P+E K V S C + L + K G + +K + +P+ + W+
Sbjct: 683 FNPYSKLFTPSERKDVITSTVCWSIMFCLLTYLSLIK-GPIVMLKLYGVPYWIFIMWLDF 859
Query: 306 FTMVHH--TAPHIPFKNSEEWNAAQAQLNGTVHCAYPQWIEILCHDINVHIPHHISPKIP 363
T +HH + +P+ +EW+ + L TV Y W+ + HDI H+ HH+ P+IP
Sbjct: 860 VTYMHHHGYSQKLPWYRGQEWDYLRGGLT-TVDRDY-GWVNNIHHDIGTHVIHHLFPQIP 1033
Query: 364 SYNLRAAHKSIQENWGKYLNE 384
Y+L A ++ + GKY E
Sbjct: 1034HYHLVEATEAAKPVLGKYYRE 1096
>TC77910 similar to GP|23345021|gb|AAN17502.1 omega-3 fatty acid desaturase
{Betula pendula}, partial (84%)
Length = 1750
Score = 102 bits (254), Expect = 3e-22
Identities = 71/276 (25%), Positives = 119/276 (42%), Gaps = 12/276 (4%)
Frame = +3
Query: 62 PAPVESAEYRKQLAERYGFEQIGEPLPDNVTLKDVITSLPKKVFEIDDVKAWKSVLISAT 121
P V+S E + + + P +L D+ ++PK + D K+ VL
Sbjct: 372 PLRVDSIEQEHEHEQEQELPEFDPGSPPPFSLADIRAAIPKHCWVKDSWKSMSYVLRDVV 551
Query: 122 S-YALGLLMISKAPWYLLPLAWAWTGTAVTGFFVIGHDCAHKSFSTNKLVEDIVGTLAFL 180
+ L W + PL WA GT FV+GHDC H SFS++ + +VG L
Sbjct: 552 IVFGLAAAAAFLNNWMVWPLYWAAQGTMFWALFVLGHDCGHGSFSSDPNLNSVVGHLLHS 731
Query: 181 PLIYPYEPWRFKHDKHHAKTNMLKEDTAWQPVWQDEFESSPFLRKAI-------IYGYGP 233
++ PY WR H HH ++ D +W P+ + F+S + +++ + Y P
Sbjct: 732 SILVPYHGWRISHRTHHQNHGHVENDESWHPLTEKIFKSLDNVTRSLRFTAPFPLLAY-P 908
Query: 234 FRCWMSIAHWLVSHF--DLKTFRPNEIKRVKISLACVFAFIAIGWPLIIYKTGIMGWIKF 291
W SHF + F P+E K + S AC A +A+ L + G + +
Sbjct: 909 VYLWARSPGKTGSHFHPSSELFLPSEKKDIITSTACWTAMVALLVGL-GFTMGPIPLLML 1085
Query: 292 WLMPWLGYHFWMSTFTMVHHTA--PHIPFKNSEEWN 325
+ +P+ + W+ T +HH +P+ +EW+
Sbjct: 1086YGVPYAIFVMWLDLVTYLHHHGHEDKLPWYRGKEWS 1193
Score = 45.8 bits (107), Expect = 3e-05
Identities = 38/156 (24%), Positives = 70/156 (44%), Gaps = 21/156 (13%)
Frame = +1
Query: 250 LKTFRPNEIKR-VKISLACVFAFIAIGWPLI--IYKTGIMGW--IKFWLMPWLGYHF--- 301
L TF P + R V IS+ V F + ++ + G++ W + W + W+ +H+
Sbjct: 907 LCTFGPEVLGRLVLISIPAVNCFFQVRKKIL*LLQLVGLLWWHCL*GWDLQWVQFHYSCF 1086
Query: 302 -------------WMSTFTMVHHTAPHIPFKNSEEWNAAQAQLNGTVHCAYPQWIEILCH 348
W T T++ A ++ E N A +++ + A + + H
Sbjct: 1087 MEFRMLFL*CGWIW*LTCTIM---AMKTNYRGIVERNGAILEVDLLLLTAIMDGLNNIHH 1257
Query: 349 DINVHIPHHISPKIPSYNLRAAHKSIQENWGKYLNE 384
DI H+ HH+ P+IP Y+L A ++ + +G+Y E
Sbjct: 1258 DIGTHVVHHLFPQIPHYHLVEATEAARPVFGEYYKE 1365
>TC77911 similar to GP|23345021|gb|AAN17502.1 omega-3 fatty acid desaturase
{Betula pendula}, partial (64%)
Length = 1079
Score = 100 bits (250), Expect = 8e-22
Identities = 72/254 (28%), Positives = 114/254 (44%), Gaps = 16/254 (6%)
Frame = +1
Query: 88 PDNVTLKDVITSLPKKVFEIDDVKAWKSV--LISATSYALGLLMISKA--PWYLLPLAWA 143
P TL D+ ++PK + D WKS+ ++ GL +++ ++ PL WA
Sbjct: 268 PPPFTLGDIRAAIPKHCWVKDP---WKSMSYVVRDVVVVFGLAVVAAYVNSLFVWPLYWA 438
Query: 144 WTGTAVTGFFVIGHDCAHKSFSTNKLVEDIVGTLAFLPLIYPYEPWRFKHDKHHAKTNML 203
GT FV+GHDC H SFS N + + G ++ PY WR H HH +
Sbjct: 439 AQGTMFWALFVLGHDCGHGSFSNNPKLNSVAGHGLHSSILVPYHGWRISHRTHHQNHGHV 618
Query: 204 KEDTAWQPVWQDEF----ESSPFLRKAIIYGY--GPFRCWMSIAHWLVSHFDLKT--FRP 255
+ D +W P+ + F S+ LR I + PF W SHFD + F P
Sbjct: 619 ENDESWHPLPEKIFRSLDNSTKMLRFTIPFPMLAYPFYLWSRSPGKKGSHFDPNSDLFVP 798
Query: 256 NEIKRVKISLAC--VFAFIAIGWPLIIYKTGIMGWIKFWLMPWLGYHFWMSTFTMVHHTA 313
NE K V S C A + +G ++ G + +K + +P++ + W+ T +HH
Sbjct: 799 NERKDVITSTVCWTAMAALLVGLGFVM---GPIQLLKLYGVPYVLFVMWLDLVTYLHHHG 969
Query: 314 --PHIPFKNSEEWN 325
+P+ +EW+
Sbjct: 970 HEDKLPWYRGQEWS 1011
>TC85814 similar to SP|P48631|FD62_SOYBN Omega-6 fatty acid desaturase
endoplasmic reticulum isozyme 2 (EC 1.14.99.-).
[Soybean], complete
Length = 1897
Score = 91.3 bits (225), Expect = 7e-19
Identities = 79/322 (24%), Positives = 138/322 (42%), Gaps = 18/322 (5%)
Frame = +2
Query: 78 YGFEQIGEPLPDNVTLKDVITSLPKKVFEIDDVKAWKSVLISATSYALGLLM-ISKAPWY 136
+ QI + +P + + VI S V+ D+ + AT Y L +S W
Sbjct: 413 FSLSQIKKAIPPHCFKRSVIHSFSYVVY---DLTIAFFLYYVATHYFQHLPRPLSFLAW- 580
Query: 137 LLPLAWAWTGTAVTGFFVIGHDCAHKSFSTNKLVEDIVGTLAFLPLIYPYEPWRFKHDKH 196
P WA G +TG +VI H+C H +FS + ++D VG + L+ PY W++ H +H
Sbjct: 581 --PAYWAIQGCILTGVWVIAHECGHHAFSDYQWLDDTVGLILHSALLVPYFSWKYSHRRH 754
Query: 197 HAKTNMLKEDTAWQP------VWQDEFESSPFLRKAIIYGYGPFRCWMSIAHWLVS---- 246
H+ T L+ D + P W ++ S+ L + + W VS
Sbjct: 755 HSNTGSLERDEVFVPKQKSSIYWYSKYLSNNPLGRVLTITITLTLGWPLYLALNVSGRPY 934
Query: 247 -----HFDL--KTFRPNEIKRVKISLACVFAFIAIGWPLIIYKTGIMGWIKFWLMPWLGY 299
HF+ + E ++ +S A + A + G + G+ + + +P L
Sbjct: 935 DRFACHFNPHGPIYSDRERLQIYVSDAGILA-VCFGLYHLAMAKGLAWVVCVYGVPLLVV 1111
Query: 300 HFWMSTFTMVHHTAPHIPFKNSEEWNAAQAQLNGTVHCAYPQWIEILCHDINVHIPHHIS 359
+ ++ T + HT P +P +S EW+ + L TV Y ++ + + H+ HH+
Sbjct: 1112NGFLVLITFLQHTHPALPHYDSSEWDWLRGAL-ATVDRDYGILNKVFHNITDTHVAHHLF 1288
Query: 360 PKIPSYNLRAAHKSIQENWGKY 381
+P Y+ A K+I+ G Y
Sbjct: 1289STMPHYHAMEATKAIKPILGDY 1354
>BE205395 homologue to SP|P48631|FD62 Omega-6 fatty acid desaturase
endoplasmic reticulum isozyme 2 (EC 1.14.99.-).
[Soybean], partial (54%)
Length = 621
Score = 41.6 bits (96), Expect = 6e-04
Identities = 24/88 (27%), Positives = 43/88 (48%)
Frame = +3
Query: 294 MPWLGYHFWMSTFTMVHHTAPHIPFKNSEEWNAAQAQLNGTVHCAYPQWIEILCHDINVH 353
+P L + ++ T + HT P +P +S EW+ + L TV Y ++ + + H
Sbjct: 294 VPLLVVNGFLVLITFLQHTHPALPHYDSSEWDWLRGAL-ATVDRDYGILNKVFHNITDTH 470
Query: 354 IPHHISPKIPSYNLRAAHKSIQENWGKY 381
+ HH+ +P Y+ A K+I+ G Y
Sbjct: 471 VAHHLFSTMPHYHAMEATKAIKPILGDY 554
>TC93794 homologue to GP|17064784|gb|AAL32546.1 temperature-sensitive
omega-3 fatty acid desaturase chloroplast precursor
{Arabidopsis thaliana}, partial (10%)
Length = 714
Score = 36.2 bits (82), Expect = 0.025
Identities = 12/26 (46%), Positives = 18/26 (69%)
Frame = +2
Query: 342 WIEILCHDINVHIPHHISPKIPSYNL 367
W+ + HDI H+ HH+ P+IP Y+L
Sbjct: 503 WLNNIHHDIGTHVVHHLFPQIPHYHL 580
>TC86624 similar to PIR|A84900|A84900 hypothetical protein At2g46210
[imported] - Arabidopsis thaliana, partial (98%)
Length = 2039
Score = 36.2 bits (82), Expect = 0.025
Identities = 52/240 (21%), Positives = 91/240 (37%), Gaps = 26/240 (10%)
Frame = +3
Query: 155 IGHDCAH----KSFSTNKLVEDIVGT-LAFLPLIYPYEPWRFKHDKHHAKTNMLKEDTAW 209
IGHD H S S NKL + + G + + + + W++ H+ HH N L D
Sbjct: 939 IGHDSGHYEVMSSRSYNKLAQILCGNCMTGISIAW----WKWTHNAHHIACNSLDYDPDL 1106
Query: 210 Q--PVWQDEFESSPFLRKAIIYGYGPFRCWMSIAHWLVSHFDLKTFRPNEIKRVKISLAC 267
Q PV+ SS F Y Y + +++ +L+S+ + + R+ + L
Sbjct: 1107 QHIPVFA---VSSRFFGSIKSYFYDRQLKFDALSRFLISYQHITFYPVLCFARLNLYLQT 1277
Query: 268 VFAFIAIGWPLIIYKTGIMGWIKFW---------LMPW-LGYHFWMSTFTM--------- 308
+ + IMG FW L W F ++ F +
Sbjct: 1278 FLLLFSPSRNVPDRLYNIMGIGVFWTWFPLLLSALPSWPERLMFVLACFVVCSIQHLQFC 1457
Query: 309 VHHTAPHIPFKNSEEWNAAQAQLNGTVHCAYPQWIEILCHDINVHIPHHISPKIPSYNLR 368
++H A ++ + Q Q GT+ W++ + + HH+ P++P LR
Sbjct: 1458 LNHFAANVYLGPPSGNDWFQKQTAGTLDITCSTWMDWFFGGLQFQLEHHLFPRLPRAQLR 1637
>CB894038
Length = 855
Score = 30.8 bits (68), Expect = 1.1
Identities = 19/72 (26%), Positives = 32/72 (44%), Gaps = 13/72 (18%)
Frame = +1
Query: 186 YEPWRFKHDKHHAKTNMLKEDTAWQPVWQDEFESSPFLRKAII------------YGYGP 233
+EP+ F +DK K ++L + + QP W +S +K + Y +G
Sbjct: 316 FEPYYFYNDKQRMKQSLLLQHKSLQPFWMIYSLTSNSWKKLYVNMPRSSPTFQLEYYHGN 495
Query: 234 FRCWM-SIAHWL 244
R +M + HWL
Sbjct: 496 HRLYMDGVCHWL 531
>TC92470 similar to GP|17065290|gb|AAL32799.1 Unknown protein {Arabidopsis
thaliana}, partial (74%)
Length = 1171
Score = 30.0 bits (66), Expect = 1.8
Identities = 10/33 (30%), Positives = 21/33 (63%), Gaps = 5/33 (15%)
Frame = +3
Query: 387 WNWRLMKTIMTVCH-----VYDKQQNYVSFDEV 414
W+W ++K IMT+CH ++ ++ N + + E+
Sbjct: 534 WDWEIVKVIMTLCHFARKGIWFQELNVIHYQEL 632
>TC90151 similar to GP|16323326|gb|AAL15376.1 At2g02370/T16F16.16
{Arabidopsis thaliana}, partial (28%)
Length = 762
Score = 29.3 bits (64), Expect = 3.1
Identities = 9/16 (56%), Positives = 12/16 (74%)
Frame = +1
Query: 285 IMGWIKFWLMPWLGYH 300
++GW FWL PWL Y+
Sbjct: 631 VVGWDDFWLWPWLCYN 678
>TC80728 similar to GP|15810469|gb|AAL07122.1 unknown protein {Arabidopsis
thaliana}, partial (87%)
Length = 1523
Score = 28.9 bits (63), Expect = 4.0
Identities = 17/55 (30%), Positives = 25/55 (44%), Gaps = 5/55 (9%)
Frame = +1
Query: 353 HIPHHISPKIPSYNLRAAHKSIQENWGKYLNEASWN--WRLMKTIMTV---CHVY 402
H+P H+ SYN H+ ++ W YL S+ +RL M + CH Y
Sbjct: 172 HLPFHLEHSRSSYNS*INHRLSRKRWFCYLGRKSFRPIFRLFNGYMEIP*WCH*Y 336
>BG451876 weakly similar to GP|15144511|gb| unknown {Lycopersicon
esculentum}, partial (7%)
Length = 671
Score = 28.5 bits (62), Expect = 5.3
Identities = 13/29 (44%), Positives = 18/29 (61%), Gaps = 4/29 (13%)
Frame = -2
Query: 336 HCAYPQ--WIEILCHDIN--VHIPHHISP 360
HC PQ W++I CHDI+ +P +SP
Sbjct: 616 HCPSPQ**WLKIRCHDISRTTFLPTAVSP 530
>TC78661 similar to PIR|T09886|T09886 hypothetical protein T22A6.60 -
Arabidopsis thaliana, partial (19%)
Length = 1116
Score = 28.5 bits (62), Expect = 5.3
Identities = 27/87 (31%), Positives = 40/87 (45%), Gaps = 2/87 (2%)
Frame = -2
Query: 233 PFR-CWMSIAHWLVSHFDLKTFRPNEIKRVKISLACVFAFIAIGWPLIIYKTGIMGWIKF 291
PFR C HWL F ++F N +K +++ L I I I + T I
Sbjct: 902 PFRTCGEFKRHWL--RFTTRSFHSNLMKSIQLHLQ-----ITITITNIPFLTNIFCSRHK 744
Query: 292 WLMPWLGYHFWMST-FTMVHHTAPHIP 317
L P HF +S ++HH++PH+P
Sbjct: 743 HLSP---PHFSLSLPIPILHHSSPHLP 672
>TC80793 similar to SP|Q9LUA9|COLE_ARATH Zinc finger protein constans-like
14. [Mouse-ear cress] {Arabidopsis thaliana}, partial
(27%)
Length = 937
Score = 28.1 bits (61), Expect = 6.9
Identities = 9/18 (50%), Positives = 12/18 (66%)
Frame = -3
Query: 180 LPLIYPYEPWRFKHDKHH 197
LP +YP+ P+R H HH
Sbjct: 896 LPYLYPFHPYRNHHTDHH 843
>BQ143784 weakly similar to SP|P33485|VNUA Probable nuclear antigen. [strain
Kaplan PRV] {Pseudorabies virus}, partial (2%)
Length = 1022
Score = 28.1 bits (61), Expect = 6.9
Identities = 8/21 (38%), Positives = 14/21 (66%)
Frame = -3
Query: 284 GIMGWIKFWLMPWLGYHFWMS 304
G+ G + FW + W G+ FW++
Sbjct: 300 GVPGCVFFWGLAWRGFAFWLA 238
>TC84227 weakly similar to GP|6682247|gb|AAF23299.1| unknown protein
{Arabidopsis thaliana}, partial (34%)
Length = 629
Score = 28.1 bits (61), Expect = 6.9
Identities = 18/62 (29%), Positives = 24/62 (38%), Gaps = 2/62 (3%)
Frame = +3
Query: 152 FFVIGHD--CAHKSFSTNKLVEDIVGTLAFLPLIYPYEPWRFKHDKHHAKTNMLKEDTAW 209
F+V+ H C H S + E I G P++ PYE H K M + W
Sbjct: 87 FWVLCHSSGCIHAWASLKYIPERIAGAAMLAPMVSPYE-------SHMTKDEMKRTWEKW 245
Query: 210 QP 211
P
Sbjct: 246 LP 251
>TC91317 similar to PIR|C96761|C96761 hypothetical protein T9L24.35
[imported] - Arabidopsis thaliana, partial (10%)
Length = 765
Score = 27.7 bits (60), Expect = 9.0
Identities = 10/14 (71%), Positives = 12/14 (85%)
Frame = -3
Query: 354 IPHHISPKIPSYNL 367
IPHH+ PKIPS +L
Sbjct: 739 IPHHVQPKIPSTSL 698
>CB894000 similar to GP|7108597|gb| K+ transporter HAK5 {Arabidopsis
thaliana}, partial (28%)
Length = 859
Score = 27.7 bits (60), Expect = 9.0
Identities = 10/34 (29%), Positives = 18/34 (52%)
Frame = +2
Query: 208 AWQPVWQDEFESSPFLRKAIIYGYGPFRCWMSIA 241
+WQP+W E E + + PF+C++ +A
Sbjct: 563 SWQPLWSSEME----------FSHHPFQCFLQLA 634
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.138 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,068,672
Number of Sequences: 36976
Number of extensions: 286350
Number of successful extensions: 1583
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 1546
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1577
length of query: 433
length of database: 9,014,727
effective HSP length: 99
effective length of query: 334
effective length of database: 5,354,103
effective search space: 1788270402
effective search space used: 1788270402
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 60 (27.7 bits)
Medicago: description of AC122160.5


