

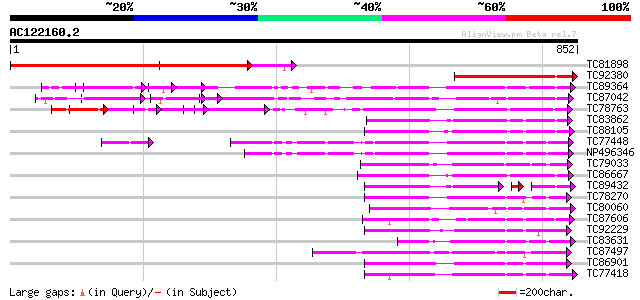
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122160.2 - phase: 0
(852 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC81898 weakly similar to GP|20260576|gb|AAM13186.1 putative rec... 717 0.0
TC92380 similar to GP|8163606|gb|AAF73754.1| receptor-like prote... 359 3e-99
TC89364 weakly similar to PIR|C96745|C96745 hypothetical protein... 225 5e-59
TC87042 similar to GP|13605827|gb|AAK32899.1 AT5g48380/MJE7_1 {A... 223 2e-58
TC78763 somatic embryogenesis receptor kinase 1 [Medicago trunca... 191 8e-49
TC83862 similar to PIR|T05270|T05270 probable serine/threonine-s... 182 4e-46
TC88105 similar to GP|21593085|gb|AAM65034.1 Putative protein ki... 181 8e-46
TC77448 SYMRK; MtSYMRK [Medicago truncatula]; nod region linked ... 181 8e-46
NP496346 NP496346|AJ418371.1|CAD10810.1 nod region linked recept... 179 3e-45
TC79033 similar to GP|21554229|gb|AAM63304.1 somatic embryogenes... 178 7e-45
TC86667 similar to GP|8778873|gb|AAF79872.1| T7N9.25 {Arabidopsi... 171 1e-42
TC89432 weakly similar to GP|9294592|dbj|BAB02873.1 protein kina... 132 4e-42
TC78270 similar to GP|16649103|gb|AAL24403.1 Unknown protein {Ar... 168 7e-42
TC80060 similar to PIR|D96574|D96574 hypothetical protein T3F20.... 167 1e-41
TC87606 similar to PIR|C84473|C84473 probable protein kinase [im... 164 1e-40
TC92229 similar to PIR|B86369|B86369 hypothetical protein AAC980... 164 1e-40
TC83631 similar to PIR|T10659|T10659 probable serine/threonine-s... 164 1e-40
TC87497 similar to GP|9759480|dbj|BAB10485.1 serine/threonine pr... 162 7e-40
TC86901 similar to GP|13937147|gb|AAK50067.1 AT5g13160/T19L5_120... 159 3e-39
TC77418 similar to GP|15146244|gb|AAK83605.1 At2g17220/T23A1.8 {... 158 1e-38
>TC81898 weakly similar to GP|20260576|gb|AAM13186.1 putative receptor-like
protein kinase {Arabidopsis thaliana}, partial (29%)
Length = 1103
Score = 717 bits (1850), Expect = 0.0
Identities = 363/364 (99%), Positives = 364/364 (99%)
Frame = +3
Query: 1 MGLGVFGSVLVLTFLFKHLASQQPNTDEFYVSEFLKKMGLTSSSKVYNFSSSVCSWKGVY 60
MGLGVFGSVLVLTFLFKHLASQQPNTDEFYVSEFLKKMGLTSSSKVYNFSSSVCSWKGVY
Sbjct: 12 MGLGVFGSVLVLTFLFKHLASQQPNTDEFYVSEFLKKMGLTSSSKVYNFSSSVCSWKGVY 191
Query: 61 CDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTLPSDFWSLTSLKSLN 120
CDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTLPSDFWSLTSLKSLN
Sbjct: 192 CDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTLPSDFWSLTSLKSLN 371
Query: 121 LSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPS 180
LSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPS
Sbjct: 372 LSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPS 551
Query: 181 GILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVSNFSRLKSIVSLN 240
GILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVSNFSRLKSIVSLN
Sbjct: 552 GILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVSNFSRLKSIVSLN 731
Query: 241 ISGNSFQGSIIEVFVLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNL 300
ISGNSFQGSIIEVFVLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNL
Sbjct: 732 ISGNSFQGSIIEVFVLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNL 911
Query: 301 NNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLS 360
NNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLG+LNALDLS
Sbjct: 912 NNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGDLNALDLS 1091
Query: 361 MNHL 364
MNHL
Sbjct: 1092MNHL 1103
Score = 67.8 bits (164), Expect = 2e-11
Identities = 64/218 (29%), Positives = 97/218 (44%), Gaps = 12/218 (5%)
Frame = +3
Query: 226 GVSNFSRLKSIVSLNISGNSFQGSIIEVFV---LKLEALDLSRNQFQGHISQVKYNWSHL 282
GV S + +V LN+SG G I + + KL +LDLS N+ S ++ + L
Sbjct: 183 GVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTLPSDF-WSLTSL 359
Query: 283 VYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVG 342
L+LS N +SG + N+ N L++ L+ N FS +
Sbjct: 360 KSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDE----------------------- 470
Query: 343 HIPDEISHLGNLNALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFILKSLPK 400
IP+ +S L +L L L N IP +LK + L ID S N LSG +P + PK
Sbjct: 471 -IPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDAFPK 647
Query: 401 MKKYNFSYNNL-------TLCASEIKPDIMKTSFFGSV 431
++ N + NN+ + S + +I SF GS+
Sbjct: 648 LRTLNLAENNIYGGVSNFSRLKSIVSLNISGNSFQGSI 761
>TC92380 similar to GP|8163606|gb|AAF73754.1| receptor-like protein kinase
{Prunus dulcis}, partial (84%)
Length = 884
Score = 359 bits (921), Expect = 3e-99
Identities = 179/184 (97%), Positives = 179/184 (97%)
Frame = +2
Query: 669 FRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKIFGSGLDEE 728
F K LGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKIFGSGLDEE
Sbjct: 26 FDTKSHLGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKIFGSGLDEE 205
Query: 729 IARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSW 788
IARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSW
Sbjct: 206 IARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSW 385
Query: 789 VRGLVRKNQTSRAIDPKICDTGSDEQIEEALKEVGYLCTADLPFKRPTMQQIVGLLKDIE 848
VRGLVRKNQTSRAIDPKICDTGSDEQIEEALK VGYLCTADLPFKRPTMQQIVGLLKDIE
Sbjct: 386 VRGLVRKNQTSRAIDPKICDTGSDEQIEEALK-VGYLCTADLPFKRPTMQQIVGLLKDIE 562
Query: 849 PTTS 852
PTTS
Sbjct: 563 PTTS 574
Score = 36.6 bits (83), Expect = 0.042
Identities = 15/23 (65%), Positives = 18/23 (78%)
Frame = +1
Query: 661 EGLLTTWRFRHKIALGTARALAF 683
EGLLTTWRFRHKIA +++ F
Sbjct: 1 EGLLTTWRFRHKIAPWNCKSIGF 69
>TC89364 weakly similar to PIR|C96745|C96745 hypothetical protein T9N14.3
[imported] - Arabidopsis thaliana, partial (45%)
Length = 1903
Score = 225 bits (574), Expect = 5e-59
Identities = 202/655 (30%), Positives = 301/655 (45%), Gaps = 14/655 (2%)
Frame = +1
Query: 209 AFPKLRTLNLAENNIYGGVSN----FSRLKSIVSLNISGNSFQGSIIEVF--VLKLEALD 262
+ P + ++L NN G VS+ + L IV +N N F G + ++ LE L
Sbjct: 58 SLPNAKIIDLGFNNFSGEVSSEIGYSTNLSEIVLMN---NKFSGKVPSEIGKLVNLEKLY 228
Query: 263 LSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFP 322
LS N F G I + L L L EN L+G I + L + L L+LA N
Sbjct: 229 LSNNNFSGDIPREIGLLKQLSTLHLEENSLTGVIPKELGHCSRLVDLNLALN-------- 384
Query: 323 KIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIP-LLKNKHLQVIDF 381
SL G+IP+ +S + +LN+L+LS N L G IP L+ L +DF
Sbjct: 385 ----------------SLSGNIPNSVSLMSSLNSLNLSRNKLTGTIPDNLEKMKLSSVDF 516
Query: 382 SHNNLSGPVPSFILKSLPKMKKYNFSYNNLTLCASEIKPDIMKTSFFGSVNSCPIAANPS 441
S N+LSG +P IL + K + N LC +I KTS + C
Sbjct: 517 SQNSLSGGIPFGIL-IIGGEKAF---VGNKELCVEQIP----KTSMNSDLKICD------ 654
Query: 442 FFKKRRDVGHR----GMKLALVLTLSLIFALAGILFLAFGCRRKNKMWEVKQGSYREEQN 497
+D GHR K L+ +++IFA A ++ C + K +++G Q
Sbjct: 655 -----KDHGHRRGVFAYKYFLLFFIAVIFAAAIVIHR---CMKNRKEKNLQKGEKEASQK 810
Query: 498 ISGPFSFQTDSTTWVADVKQATSVPVVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFG 557
W KQA+ V I + S+ L+ G G
Sbjct: 811 -------------W----KQASFHQVDIDADEV------------SHLGDDNLIGYGGTG 903
Query: 558 PVYRGFLPGN-IHVAVKVLVVGSTLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIA 616
VYR L I VAVK L G + + A E+E L +I+H N++ L + G +
Sbjct: 904 KVYRVKLKKTGIVVAVKQLEKGYGV--KILAAEMEILAKIRHKNILKLYACFLKGGSNLL 1077
Query: 617 IYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALG 676
+++YM NGNL L+ + V E + W R+KIALG
Sbjct: 1078VFEYMPNGNLFQALH------------------------REVKDEMVTFDWNQRYKIALG 1185
Query: 677 TARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKIF-GSGLDEEIARGSPG 735
A+ + +LHH CSPP+IHR +K+S++ LD + E +++DFG+A+ S + + G+ G
Sbjct: 1186GAKGICYLHHDCSPPVIHRDIKSSNILLDANYEAKIADFGVARFAEKSQMGYSVFAGTHG 1365
Query: 736 YVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRK 795
Y+ PE + T KSDVY FGVVL EL++G++PV ++Y EA +V WV +
Sbjct: 1366YIAPELAYT--TEITEKSDVYSFGVVLLELVSGREPVEEEY---GEAKDIVHWVMSNLND 1530
Query: 796 NQTSRAI-DPKICDTGSDEQIEEALKEVGYLCTADLPFKRPTMQQIVGLLKDIEP 849
++ +I D ++ T E + + LK +G CT LP RPTM+ +V +L D EP
Sbjct: 1531RESVLSILDGRVASTHCVEDMIKVLK-IGIKCTTKLPTLRPTMRDVVKMLVDSEP 1692
Score = 78.2 bits (191), Expect = 1e-14
Identities = 48/151 (31%), Positives = 73/151 (47%)
Frame = +1
Query: 100 NNKITTLPSDFWSLTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALS 159
NN + S+ T+L + L +N SG + + IG LE LS N+FS +IP +
Sbjct: 94 NNFSGEVSSEIGYSTNLSEIVLMNNKFSGKVPSEIGKLVNLEKLYLSNNNFSGDIPREIG 273
Query: 160 SLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLA 219
L L L L+ N IP + C LV ++L+ N LSG +P+ L +LNL+
Sbjct: 274 LLKQLSTLHLEENSLTGVIPKELGHCSRLVDLNLALNSLSGNIPNSV-SLMSSLNSLNLS 450
Query: 220 ENNIYGGVSNFSRLKSIVSLNISGNSFQGSI 250
N + G + + + S++ S NS G I
Sbjct: 451 RNKLTGTIPDNLEKMKLSSVDFSQNSLSGGI 543
Score = 77.8 bits (190), Expect = 2e-14
Identities = 55/159 (34%), Positives = 82/159 (50%), Gaps = 1/159 (0%)
Frame = +1
Query: 48 NFSSSVCSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TL 106
NFS V S G +N +V +N +G +P + IGKL L L LSNN + +
Sbjct: 97 NFSGEVSSEIGY--STNLSEIVLMNNK---FSGKVP-SEIGKLVNLEKLYLSNNNFSGDI 258
Query: 107 PSDFWSLTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKV 166
P + L L +L+L N ++G + +G+ L + +L+ NS S IP ++S + SL
Sbjct: 259 PREIGLLKQLSTLHLEENSLTGVIPKELGHCSRLVDLNLALNSLSGNIPNSVSLMSSLNS 438
Query: 167 LKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHG 205
L L N +IP + K + L S+D S N LSG +P G
Sbjct: 439 LNLSRNKLTGTIPDNLEKMK-LSSVDFSQNSLSGGIPFG 552
Score = 71.2 bits (173), Expect = 2e-12
Identities = 58/189 (30%), Positives = 86/189 (44%), Gaps = 3/189 (1%)
Frame = +1
Query: 111 WSLTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLD 170
WSL + K ++L N+ SG +++ IG L L N FS ++P + LV+L+ L L
Sbjct: 55 WSLPNAKIIDLGFNNFSGEVSSEIGYSTNLSEIVLMNNKFSGKVPSEIGKLVNLEKLYL- 231
Query: 171 HNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SN 229
S+N SG +P G +L TL+L EN++ G +
Sbjct: 232 -----------------------SNNNFSGDIPREIG-LLKQLSTLHLEENSLTGVIPKE 339
Query: 230 FSRLKSIVSLNISGNSFQGSIIEVFVL--KLEALDLSRNQFQGHISQVKYNWSHLVYLDL 287
+V LN++ NS G+I L L +L+LSRN+ G I L +D
Sbjct: 340 LGHCSRLVDLNLALNSLSGNIPNSVSLMSSLNSLNLSRNKLTGTIPD-NLEKMKLSSVDF 516
Query: 288 SENQLSGEI 296
S+N LSG I
Sbjct: 517 SQNSLSGGI 543
>TC87042 similar to GP|13605827|gb|AAK32899.1 AT5g48380/MJE7_1 {Arabidopsis
thaliana}, partial (51%)
Length = 2206
Score = 223 bits (569), Expect = 2e-58
Identities = 172/573 (30%), Positives = 266/573 (46%), Gaps = 10/573 (1%)
Frame = +2
Query: 285 LDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLG-LEYLNLSKTSLVGH 343
L LS L G+ + + N ++ L L+ N S I LL + L+LS G
Sbjct: 395 LKLSNMGLKGQFPRGIVNCSSMTGLDLSVNDLSGTIPGDISTLLKFVTSLDLSSNEFSGE 574
Query: 344 IPDEISHLGNLNALDLSMNHLDGKIPLLKNK--HLQVIDFSHNNLSGPVPSFILKSLPKM 401
IP +++ LN L LS N L G+IPLL ++ D S+N L+G VP+F
Sbjct: 575 IPVSLANCTYLNVLKLSQNQLTGQIPLLLGTLDRIKTFDVSNNLLTGQVPNFTAGG---- 742
Query: 402 KKYNFSY-NNLTLCASEIKPDIMKTSFFGSVNSCPIAANPSFFKKRRDVGHRGMKLALVL 460
K + +Y NN LC + K + N+ IA A V
Sbjct: 743 -KVDVNYANNQGLCGQP-SLGVCKATASSKSNTAVIAG------------------AAVG 862
Query: 461 TLSLIFALAGILFLAFGCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADVKQATS 520
++L G+ F V++ +YR+++ + W +K
Sbjct: 863 AVTLAALGLGVFMFFF----------VRRSAYRKKEE-------DPEGNKWARSLKGTKG 991
Query: 521 VPVVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGST 580
+ V +FEK + + +DL+ AT+NF ++ G+ G VY+ L VK L S
Sbjct: 992 IKVSLFEKSISKMKLSDLMKATNNFSNINIIGTGRTGTVYKATLEDGTAFMVKRLQE-SQ 1168
Query: 581 LTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQST 640
+++E E+ LG +KH NLVPL G+CVA +R+ ++ M NG L + L+ P + T
Sbjct: 1169HSEKEFMSEMATLGTVKHRNLVPLLGFCVAKKERLLVFKNMPNGMLHDQLH--PAAGECT 1342
Query: 641 DDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKAS 700
DW + R KIA+G A+ A+LHH C+P IIHR + +
Sbjct: 1343LDWPS------------------------RLKIAIGAAKGFAWLHHSCNPRIIHRNISSK 1450
Query: 701 SVYLDYDLEPRLSDFGLAKIFGSGLDEEIAR------GSPGYVPPEFSQPEFESPTPKSD 754
+ LD D EP++SDFGLA++ + LD ++ G GYV PE+++ TPK D
Sbjct: 1451CILLDADFEPKISDFGLARLM-NPLDTHLSTFVNGEFGDFGYVAPEYTKTLV--ATPKGD 1621
Query: 755 VYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQ 814
V+ FG VL EL+TG++P + LV W+ L ++ AID + + G D +
Sbjct: 1622VFSFGTVLLELVTGERPANVAKAPETFKGNLVEWITELSSNSKLHDAIDESLLNKGDDNE 1801
Query: 815 IEEALKEVGYLCTADLPFKRPTMQQIVGLLKDI 847
+ + LK V C ++P +RPTM ++ L+ I
Sbjct: 1802LFQFLK-VACNCVTEVPKERPTMFEVYQFLRAI 1897
Score = 66.6 bits (161), Expect = 4e-11
Identities = 53/173 (30%), Positives = 82/173 (46%), Gaps = 8/173 (4%)
Frame = +2
Query: 40 LTSSSKVYNFSSS----VCSWKGVYC---DSNKEHVVELNLSGIGLTGPIPDTTIGKLNK 92
LTSS +NF++ +C + GV C D NK V+ L LS +GL G P +
Sbjct: 296 LTSS---WNFNNKTEGFICRFNGVECWHPDENK--VLNLKLSNMGLKGQFPRGIV----- 445
Query: 93 LHSLDLSNNKITTLPSDFWSLTSLKSLNLSSNHISGSLTNNIGNF-GLLENFDLSKNSFS 151
+ +S+ L+LS N +SG++ +I + + DLS N FS
Sbjct: 446 -------------------NCSSMTGLDLSVNDLSGTIPGDISTLLKFVTSLDLSSNEFS 568
Query: 152 DEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPH 204
EIP +L++ L VLKL N IP + + + D+S+N L+G +P+
Sbjct: 569 GEIPVSLANCTYLNVLKLSQNQLTGQIPLLLGTLDRIKTFDVSNNLLTGQVPN 727
Score = 57.0 bits (136), Expect = 3e-08
Identities = 54/193 (27%), Positives = 87/193 (44%), Gaps = 4/193 (2%)
Frame = +2
Query: 108 SDFWSLTSLK-SLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKV 166
+D + L S+K S+ +N+++ S N G + F+ + DE + +++LK+
Sbjct: 239 TDIFCLKSIKNSIQDPNNYLTSSWNFNNKTEGFICRFNGVECWHPDE-----NKVLNLKL 403
Query: 167 LKLDHNMFVRS-IPSGILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYG 225
NM ++ P GI+ C S+ +DLS N LSGT+P G
Sbjct: 404 ----SNMGLKGQFPRGIVNCSSMTGLDLSVNDLSGTIP---------------------G 508
Query: 226 GVSNFSRLKSIVSLNISGNSFQGSIIEVFV--LKLEALDLSRNQFQGHISQVKYNWSHLV 283
+S LK + SL++S N F G I L L LS+NQ G I + +
Sbjct: 509 DISTL--LKFVTSLDLSSNEFSGEIPVSLANCTYLNVLKLSQNQLTGQIPLLLGTLDRIK 682
Query: 284 YLDLSENQLSGEI 296
D+S N L+G++
Sbjct: 683 TFDVSNNLLTGQV 721
Score = 43.9 bits (102), Expect = 3e-04
Identities = 36/115 (31%), Positives = 57/115 (49%), Gaps = 7/115 (6%)
Frame = +2
Query: 212 KLRTLNLAENNIYG----GVSNFSRLKSIVSLNISGNSFQGSI---IEVFVLKLEALDLS 264
K+ L L+ + G G+ N S S+ L++S N G+I I + + +LDLS
Sbjct: 383 KVLNLKLSNMGLKGQFPRGIVNCS---SMTGLDLSVNDLSGTIPGDISTLLKFVTSLDLS 553
Query: 265 RNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQ 319
N+F G I N ++L L LS+NQL+G+I L +K ++ N + Q
Sbjct: 554 SNEFSGEIPVSLANCTYLNVLKLSQNQLTGQIPLLLGTLDRIKTFDVSNNLLTGQ 718
>TC78763 somatic embryogenesis receptor kinase 1 [Medicago truncatula]
Length = 2737
Score = 191 bits (486), Expect = 8e-49
Identities = 170/582 (29%), Positives = 253/582 (43%), Gaps = 14/582 (2%)
Frame = +3
Query: 278 NWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSK 337
N + ++ +DL LSG + L NL++L L N + + L L L+L
Sbjct: 627 NDNSVIRVDLGNAALSGTLVPQLGQLKNLQYLELYSNNITGPIPSDLGNLTNLVSLDLYL 806
Query: 338 TSLVGHIPDEISHLGNLNALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFIL 395
G IPD + L L L L+ N L G IP L LQV+D S+N LSG VP
Sbjct: 807 NRFNGPIPDSLGKLSKLRFLRLNNNSLMGPIPMSLTNISALQVLDLSNNQLSGVVPD--N 980
Query: 396 KSLPKMKKYNFSYNNLTLCASEIKPDIMKTSFFGSVNSCPIAANPSF-----FKKRRDVG 450
S +F+ NNL LC G V P +P F F +
Sbjct: 981 GSFSLFTPISFA-NNLNLC--------------GPVTGHPCPGSPPFSPPPPFVPPPPIS 1115
Query: 451 HRGMKLALVLTLSLIFALAGILF----LAFGCRRKNKMWEVKQGSYREEQNISGPFSFQT 506
G A + A A +LF +AF R+ K E F F
Sbjct: 1116 APGSGGATGAIAGGVAAGAALLFAAPAIAFAWWRRRKPQE---------------FFF-- 1244
Query: 507 DSTTWVADVKQATSVPVVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPG 566
DV V + + L + +L AT F +L G FG VY+G L
Sbjct: 1245 -------DVPAEEDPEVHLGQ--LKRFSLRELQVATDTFSNKNILGRGGFGKVYKGRLAD 1397
Query: 567 NIHVAVKVLVVGSTLTDE-EAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGN 625
VAVK L T E + E+E + H NL+ L G+C+ +R+ +Y YM NG+
Sbjct: 1398 GSLVAVKRLKEERTPGGELQFQTEVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGS 1577
Query: 626 LQNLLYDLPLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLH 685
+ + L + P + D W R +IALG+AR L++LH
Sbjct: 1578 VASCLRERPPHQEPLD-------------------------WPTRKRIALGSARGLSYLH 1682
Query: 686 HGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKI--FGSGLDEEIARGSPGYVPPEFSQ 743
C P IIHR VKA+++ LD + E + DFGLAK+ + RG+ G++ PE+
Sbjct: 1683 DHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTHVTTAVRGTIGHIAPEYLS 1862
Query: 744 PEFESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAID 803
S K+DV+ +G++L EL+TG++ + + L+ WV+GL+++ + +D
Sbjct: 1863 TGKSS--EKTDVFGYGIMLLELITGQRAFDLARLANDDDVMLLDWVKGLLKEKKLEMLVD 2036
Query: 804 PKICDTGSDEQIEEALKEVGYLCTADLPFKRPTMQQIVGLLK 845
P + + ++E+ L +V LCT P RP M +V +L+
Sbjct: 2037 PDLKTNYIEAEVEQ-LIQVALLCTQGSPMDRPKMSDVVRMLE 2159
Score = 74.3 bits (181), Expect = 2e-13
Identities = 47/140 (33%), Positives = 78/140 (55%), Gaps = 2/140 (1%)
Frame = +3
Query: 91 NKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNS 149
N + +DL N ++ TL L +L+ L L SN+I+G + +++GN L + DL N
Sbjct: 633 NSVIRVDLGNAALSGTLVPQLGQLKNLQYLELYSNNITGPIPSDLGNLTNLVSLDLYLNR 812
Query: 150 FSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDA 209
F+ IP++L L L+ L+L++N + IP + +L +DLS+NQLSG +P +
Sbjct: 813 FNGPIPDSLGKLSKLRFLRLNNNSLMGPIPMSLTNISALQVLDLSNNQLSGVVPD--NGS 986
Query: 210 FPKLRTLNLAEN-NIYGGVS 228
F ++ A N N+ G V+
Sbjct: 987 FSLFTPISFANNLNLCGPVT 1046
Score = 63.9 bits (154), Expect = 2e-10
Identities = 46/132 (34%), Positives = 66/132 (49%), Gaps = 2/132 (1%)
Frame = +3
Query: 261 LDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQK 320
+DL G + +L YL+L N ++G I +L N NL L L NRF+
Sbjct: 648 VDLGNAALSGTLVPQLGQLKNLQYLELYSNNITGPIPSDLGNLTNLVSLDLYLNRFNGPI 827
Query: 321 FPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIPLLKNKHL-QVI 379
+ L L +L L+ SL+G IP ++++ L LDLS N L G +P + L I
Sbjct: 828 PDSLGKLSKLRFLRLNNNSLMGPIPMSLTNISALQVLDLSNNQLSGVVPDNGSFSLFTPI 1007
Query: 380 DFSHN-NLSGPV 390
F++N NL GPV
Sbjct: 1008SFANNLNLCGPV 1043
Score = 59.3 bits (142), Expect = 6e-09
Identities = 45/116 (38%), Positives = 60/116 (50%), Gaps = 3/116 (2%)
Frame = +3
Query: 187 SLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLNISGNS 245
S++ +DL + LSGTL G L+ L L NNI G + S+ L ++VSL++ N
Sbjct: 636 SVIRVDLGNAALSGTLVPQLGQ-LKNLQYLELYSNNITGPIPSDLGNLTNLVSLDLYLNR 812
Query: 246 FQGSIIEVF--VLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQN 299
F G I + + KL L L+ N G I N S L LDLS NQLSG + N
Sbjct: 813 FNGPIPDSLGKLSKLRFLRLNNNSLMGPIPMSLTNISALQVLDLSNNQLSGVVPDN 980
Score = 50.1 bits (118), Expect = 4e-06
Identities = 31/86 (36%), Positives = 52/86 (60%), Gaps = 1/86 (1%)
Frame = +3
Query: 64 NKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLS 122
N ++V L+L GPIPD+ +GKL+KL L L+NN + +P ++++L+ L+LS
Sbjct: 771 NLTNLVSLDLYLNRFNGPIPDS-LGKLSKLRFLRLNNNSLMGPIPMSLTNISALQVLDLS 947
Query: 123 SNHISGSLTNNIGNFGLLENFDLSKN 148
+N +SG + +N G+F L + N
Sbjct: 948 NNQLSGVVPDN-GSFSLFTPISFANN 1022
Score = 31.6 bits (70), Expect = 1.3
Identities = 19/64 (29%), Positives = 37/64 (57%), Gaps = 1/64 (1%)
Frame = +3
Query: 71 LNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTLPSDFWSLTSLKSLNLSSN-HISGS 129
L L+ L GPIP ++ ++ L LDLSNN+++ + D S + ++ ++N ++ G
Sbjct: 864 LRLNNNSLMGPIP-MSLTNISALQVLDLSNNQLSGVVPDNGSFSLFTPISFANNLNLCGP 1040
Query: 130 LTNN 133
+T +
Sbjct: 1041VTGH 1052
>TC83862 similar to PIR|T05270|T05270 probable serine/threonine-specific
protein kinase (EC 2.7.1.-) T4L20.80 - Arabidopsis
thaliana, partial (60%)
Length = 948
Score = 182 bits (463), Expect = 4e-46
Identities = 115/311 (36%), Positives = 167/311 (52%), Gaps = 2/311 (0%)
Frame = +1
Query: 537 DLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARELEFLGRI 596
+L +AT F G+++ E +G VYRG L VAVK L+ ++E E+E +G++
Sbjct: 25 ELENATDGFAEGSVIGERGYGIVYRGILQDGSIVAVKNLLNNKGQAEKEFKVEVEAIGKV 204
Query: 597 KHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQ 656
+H NLV L GYC G +R+ +Y+Y++NGNL+ L+
Sbjct: 205 RHKNLVGLVGYCAEGAKRMLVYEYVDNGNLEQWLHG------------------------ 312
Query: 657 NVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFG 716
+VG L TW R KIA+GTA+ LA+LH G P ++HR VK+S++ LD ++SDFG
Sbjct: 313 DVGPVSPL-TWDIRMKIAVGTAKGLAYLHEGLEPKVVHRDVKSSNILLDKKWHAKVSDFG 489
Query: 717 LAKIFGSGLDEEIAR--GSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGD 774
LAK+ GSG R G+ GYV PE++ + SDVY FG++L EL+TG+ P+
Sbjct: 490 LAKLLGSGKSYVTTRVMGTFGYVSPEYASTGMLN--EGSDVYSFGILLMELVTGRSPI-- 657
Query: 775 DYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKEVGYLCTADLPFKR 834
DY+ LV W +G+V + +DP I S ++ AL V C KR
Sbjct: 658 DYSRAPAEMNLVDWFKGMVASRRGEELVDPLIEIQPSPRSLKRALL-VCLRCIDLDANKR 834
Query: 835 PTMQQIVGLLK 845
P M QIV +L+
Sbjct: 835 PKMGQIVHMLE 867
>TC88105 similar to GP|21593085|gb|AAM65034.1 Putative protein kinase
{Arabidopsis thaliana}, partial (72%)
Length = 2379
Score = 181 bits (460), Expect = 8e-46
Identities = 110/317 (34%), Positives = 162/317 (50%), Gaps = 2/317 (0%)
Frame = +2
Query: 534 TFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARELEFL 593
T DL AT+ F + ++ EG +G VY+G L VA+K L+ ++E E+E +
Sbjct: 692 TLRDLELATNKFSKDNIIGEGGYGVVYQGQLINGNPVAIKKLLNNLGQAEKEFRVEVEAI 871
Query: 594 GRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADN 653
G ++H NLV L G+C+ G R+ IY+Y+ NGNL+ L+
Sbjct: 872 GHVRHKNLVRLLGFCIEGTHRLLIYEYVNNGNLEQWLH---------------------G 988
Query: 654 GIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLS 713
++ G TW R KI LGTA+ALA+LH P ++HR +K+S++ +D D ++S
Sbjct: 989 AMRQYG----YLTWDARIKILLGTAKALAYLHEAIEPKVVHRDIKSSNILIDDDFNAKIS 1156
Query: 714 DFGLAKIFGSGLDEEIAR--GSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKP 771
DFGLAK+ G+G R G+ GYV PE++ + KSDVY FGV+L E +TG+ P
Sbjct: 1157DFGLAKLLGAGKSHITTRVMGTFGYVAPEYANSGLLN--EKSDVYSFGVLLLEAITGRDP 1330
Query: 772 VGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKEVGYLCTADLP 831
V DY LV W++ +V +DP I S ++ L C
Sbjct: 1331V--DYNRSAAEVNLVDWLKMMVGNRHAEEVVDPNIETRPSTSALKRVLL-TALRCVDPDS 1501
Query: 832 FKRPTMQQIVGLLKDIE 848
KRP M Q+V +L+ E
Sbjct: 1502EKRPKMSQVVRMLESEE 1552
>TC77448 SYMRK; MtSYMRK [Medicago truncatula]; nod region linked receptor
kinase [Medicago truncatula]
Length = 3568
Score = 181 bits (460), Expect = 8e-46
Identities = 160/519 (30%), Positives = 237/519 (44%), Gaps = 5/519 (0%)
Frame = +1
Query: 333 LNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIPLLKNKHLQV-IDFSHNNLSGPVP 391
L+LS +L G IP ++ + NL L+LS N D P L + +D S+N+LSG +P
Sbjct: 1606 LDLSSNNLKGAIPSIVTKMTNLQILNLSHNQFDMLFPSFPPSSLLISLDLSYNDLSGWLP 1785
Query: 392 SFILKSLPKMKKYNFSYNNLTLCASEIKPDIMKTSFFGSVNSCPIAANPSFFKKRRDVGH 451
I+ SLP +K F N S D K +NS I + K ++
Sbjct: 1786 ESII-SLPHLKSLYFGCN-----PSMSDEDTTK------LNSSLINTDYGRCKAKKPKFG 1929
Query: 452 RGMKLALVLTLSLIFALA-GILFLAFGCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTT 510
+ + + + SL+ LA GILF CR ++K ++ + FS +
Sbjct: 1930 QVFVIGAITSGSLLITLAVGILFF---CRYRHKSITLEGFGKTYPMATNIIFSLPSKDDF 2100
Query: 511 WVADVKQATSVPVVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHV 570
++ V KP T + AT + TL+ EG FG VYRG L V
Sbjct: 2101 FIKSVSV----------KPF---TLEYIEQATEQYK--TLIGEGGFGSVYRGTLDDGQEV 2235
Query: 571 AVKVLVVGSTLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLL 630
AVKV ST E EL L I+H NLVPL GYC DQ+I +Y +M NG+L + L
Sbjct: 2236 AVKVRSSTSTQGTREFDNELNLLSAIQHENLVPLLGYCNEYDQQILVYPFMSNGSLLDRL 2415
Query: 631 YDLPLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSP 690
Y S+ + W R IALG AR LA+LH
Sbjct: 2416 Y-------------------------GEASKRKILDWPTRLSIALGAARGLAYLHTFPGR 2520
Query: 691 PIIHRAVKASSVYLDYDLEPRLSDFGLAKIFGSGLDEEIA---RGSPGYVPPEFSQPEFE 747
+IHR VK+S++ LD + +++DFG +K D ++ RG+ GY+ PE+ + +
Sbjct: 2521 SVIHRDVKSSNILLDQSMCAKVADFGFSKYAPQEGDSYVSLEVRGTAGYLDPEYYKT--Q 2694
Query: 748 SPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKIC 807
+ KSDV+ FGVVL E+++G++P+ + + +LV W + +R ++ +DP I
Sbjct: 2695 QLSEKSDVFSFGVVLLEIVSGREPL--NIKRPRIEWSLVEWAKPYIRASKVDEIVDPGI- 2865
Query: 808 DTGSDEQIEEALKEVGYLCTADLPFKRPTMQQIVGLLKD 846
G + + EV C RP M IV L+D
Sbjct: 2866 KGGYHAEALWRVVEVALQCLEPYSTYRPCMVDIVRELED 2982
Score = 49.3 bits (116), Expect = 6e-06
Identities = 28/78 (35%), Positives = 42/78 (52%)
Frame = +1
Query: 139 LLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQL 198
++ DLS N+ IP ++ + +L++L L HN F PS L+S+DLS N L
Sbjct: 1594 IITKLDLSSNNLKGAIPSIVTKMTNLQILNLSHNQFDMLFPS-FPPSSLLISLDLSYNDL 1770
Query: 199 SGTLPHGFGDAFPKLRTL 216
SG LP + P L++L
Sbjct: 1771 SGWLPESI-ISLPHLKSL 1821
Score = 41.2 bits (95), Expect = 0.002
Identities = 36/113 (31%), Positives = 51/113 (44%), Gaps = 1/113 (0%)
Frame = +1
Query: 56 WKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLT 114
WKG+ CD D+T + + LDLS+N + +PS +T
Sbjct: 1555 WKGITCD---------------------DSTGSSI--ITKLDLSSNNLKGAIPSIVTKMT 1665
Query: 115 SLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVL 167
+L+ LNLS N L + LL + DLS N S +PE++ SL LK L
Sbjct: 1666 NLQILNLSHNQFD-MLFPSFPPSSLLISLDLSYNDLSGWLPESIISLPHLKSL 1821
Score = 40.8 bits (94), Expect = 0.002
Identities = 28/81 (34%), Positives = 45/81 (54%), Gaps = 2/81 (2%)
Frame = +1
Query: 236 IVSLNISGNSFQGSIIEVF--VLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLS 293
I L++S N+ +G+I + + L+ L+LS NQF + S L+ LDLS N LS
Sbjct: 1597 ITKLDLSSNNLKGAIPSIVTKMTNLQILNLSHNQFDMLFPSFPPS-SLLISLDLSYNDLS 1773
Query: 294 GEIFQNLNNSMNLKHLSLACN 314
G + +++ + +LK L CN
Sbjct: 1774 GWLPESIISLPHLKSLYFGCN 1836
Score = 39.7 bits (91), Expect = 0.005
Identities = 29/85 (34%), Positives = 42/85 (49%), Gaps = 2/85 (2%)
Frame = +1
Query: 280 SHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSR--QKFPKIEMLLGLEYLNLSK 337
S + LDLS N L G I + NL+ L+L+ N+F FP +L+ L+ LS
Sbjct: 1591 SIITKLDLSSNNLKGAIPSIVTKMTNLQILNLSHNQFDMLFPSFPPSSLLISLD---LSY 1761
Query: 338 TSLVGHIPDEISHLGNLNALDLSMN 362
L G +P+ I L +L +L N
Sbjct: 1762 NDLSGWLPESIISLPHLKSLYFGCN 1836
>NP496346 NP496346|AJ418371.1|CAD10810.1 nod region linked receptor kinase
[Medicago truncatula]
Length = 2706
Score = 179 bits (455), Expect = 3e-45
Identities = 156/499 (31%), Positives = 231/499 (46%), Gaps = 6/499 (1%)
Frame = +1
Query: 354 LNALDLSMNHLDGKIPLLKNK--HLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNL 411
+ LDLS N+L G IP + K +LQ++D S+N+LSG +P I+ SLP +K F N
Sbjct: 1222 ITKLDLSSNNLKGAIPSIVTKMTNLQILDLSYNDLSGWLPESII-SLPHLKSLYFGCN-- 1392
Query: 412 TLCASEIKPDIMKTSFFGSVNSCPIAANPSFFKKRRDVGHRGMKLALVLTLSLIFALA-G 470
S D K +NS I + K ++ + + + + SL+ LA G
Sbjct: 1393 ---PSMSDEDTTK------LNSSLINTDYGRCKAKKPKFGQVFVIGAITSGSLLITLAVG 1545
Query: 471 ILFLAFGCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADVKQATSVPVVIFEKPL 530
ILF CR ++K ++ + FS + ++ V KP
Sbjct: 1546 ILFF---CRYRHKSITLEGFGKTYPMATNIIFSLPSKDDFFIKSVSV----------KPF 1686
Query: 531 LNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAAREL 590
T + AT + TL+ EG FG VYRG L VAVKV ST E EL
Sbjct: 1687 ---TLEYIEQATEQYK--TLIGEGGFGSVYRGTLDDGQEVAVKVRSSTSTQGTREFDNEL 1851
Query: 591 EFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEE 650
L I+H NLVPL GYC DQ+I +Y +M NG+L + LY
Sbjct: 1852 NLLSAIQHENLVPLLGYCNEYDQQILVYPFMSNGSLLDRLY------------------- 1974
Query: 651 ADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEP 710
S+ + W R IALG AR LA+LH +IHR VK+S++ LD +
Sbjct: 1975 ------GEASKRKILDWPTRLSIALGAARGLAYLHTFPGRSVIHRDVKSSNILLDQSMCA 2136
Query: 711 RLSDFGLAKIFGSGLDEEIA---RGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLT 767
+++DFG +K D ++ RG+ GY+ PE+ + + + KSDV+ FGVVL E+++
Sbjct: 2137 KVADFGFSKYAPQEGDSYVSLEVRGTAGYLDPEYYKT--QQLSEKSDVFSFGVVLLEIVS 2310
Query: 768 GKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKEVGYLCT 827
G++P+ + + +LV W + +R ++ +DP I G + + EV C
Sbjct: 2311 GREPL--NIKRPRIEWSLVEWAKPYIRASKVDEIVDPGI-KGGYHAEALWRVVEVALQCL 2481
Query: 828 ADLPFKRPTMQQIVGLLKD 846
RP M IV L+D
Sbjct: 2482 EPYSTYRPCMVDIVRELED 2538
Score = 38.5 bits (88), Expect = 0.011
Identities = 21/54 (38%), Positives = 31/54 (56%)
Frame = +1
Query: 261 LDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACN 314
LDLS N +G I + ++L LDLS N LSG + +++ + +LK L CN
Sbjct: 1231 LDLSSNNLKGAIPSIVTKMTNLQILDLSYNDLSGWLPESIISLPHLKSLYFGCN 1392
Score = 35.8 bits (81), Expect = 0.071
Identities = 19/52 (36%), Positives = 31/52 (59%)
Frame = +1
Query: 116 LKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVL 167
+ L+LSSN++ G++ + + L+ DLS N S +PE++ SL LK L
Sbjct: 1222 ITKLDLSSNNLKGAIPSIVTKMTNLQILDLSYNDLSGWLPESIISLPHLKSL 1377
Score = 34.7 bits (78), Expect = 0.16
Identities = 46/167 (27%), Positives = 69/167 (40%), Gaps = 11/167 (6%)
Frame = +1
Query: 56 WKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLT 114
WKG+ CD D+T + + LDLS+N + +PS +T
Sbjct: 1180 WKGITCD---------------------DSTGSSI--ITKLDLSSNNLKGAIPSIVTKMT 1290
Query: 115 SLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKN-SFSDEIPEAL-SSLVS-------LK 165
+L+ L+LS N +SG L +I + L++ N S SDE L SSL++ K
Sbjct: 1291 NLQILDLSYNDLSGWLPESIISLPHLKSLYFGCNPSMSDEDTTKLNSSLINTDYGRCKAK 1470
Query: 166 VLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLP-HGFGDAFP 211
K + +I SG L V I ++ GFG +P
Sbjct: 1471 KPKFGQVFVIGAITSGSLLITLAVGILFFCRYRHKSITLEGFGKTYP 1611
Score = 31.6 bits (70), Expect = 1.3
Identities = 20/50 (40%), Positives = 27/50 (54%)
Frame = +1
Query: 167 LKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTL 216
L L N +IPS + K +L +DLS N LSG LP + P L++L
Sbjct: 1231 LDLSSNNLKGAIPSIVTKMTNLQILDLSYNDLSGWLPESI-ISLPHLKSL 1377
>TC79033 similar to GP|21554229|gb|AAM63304.1 somatic embryogenesis
receptor-like kinase putative {Arabidopsis thaliana},
partial (77%)
Length = 1762
Score = 178 bits (452), Expect = 7e-45
Identities = 118/324 (36%), Positives = 166/324 (50%), Gaps = 5/324 (1%)
Frame = +1
Query: 527 EKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEA 586
+ P + +L SAT+NF+ L EG FG VY G L +AVK L V S D E
Sbjct: 409 QPPWRVFSLKELHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKVWSNKADMEF 588
Query: 587 ARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTD 646
A E+E L R++H NL+ L GYC G +R+ +YDYM N +L + L+ G ST+
Sbjct: 589 AVEVEILARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLH----GQHSTES---- 744
Query: 647 TWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDY 706
L W R IA+G+A + +LH +P IIHR VKAS+V LD
Sbjct: 745 -----------------LLDWNRRMNIAIGSAEGIVYLHVQATPHIIHRDVKASNVLLDS 873
Query: 707 DLEPRLSDFGLAKIFGSGLDEEIAR--GSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFE 764
D + R++DFG AK+ G R G+ GY+ PE++ DVY FG++L E
Sbjct: 874 DFQARVADFGFAKLIPDGATHVTTRVKGTLGYLAPEYAM--LGKANESCDVYSFGILLLE 1047
Query: 765 LLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKE--- 821
L +GKKP+ + K A + W L + + S DP++ + + +EE LK
Sbjct: 1048LASGKKPLEKLSSSVKRA--INDWALPLACEKKFSELADPRL----NGDYVEEELKRVIL 1209
Query: 822 VGYLCTADLPFKRPTMQQIVGLLK 845
V +C + P KRPTM ++V LLK
Sbjct: 1210VALICAQNQPEKRPTMVEVVELLK 1281
>TC86667 similar to GP|8778873|gb|AAF79872.1| T7N9.25 {Arabidopsis
thaliana}, partial (52%)
Length = 1443
Score = 171 bits (433), Expect = 1e-42
Identities = 109/328 (33%), Positives = 165/328 (50%), Gaps = 3/328 (0%)
Frame = +3
Query: 523 VVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLT 582
++ F LL + DL++AT+NF +L + G YR LP +AVK L +
Sbjct: 93 LIFFRNRLLR*SXGDLMAATNNFSNENVLITTRTGATYRADLPDGSTLAVKRLS-SCKIG 269
Query: 583 DEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDD 642
+++ E+ LG+++HPNL PL GYCV ++++ +Y +M NG L +LL+
Sbjct: 270 EKQFRMEMNRLGQVRHPNLAPLLGYCVVEEEKLLVYKHMSNGTLYSLLH----------- 416
Query: 643 WSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSV 702
+N G + W R +I LG AR LA+LHHGC PPII + + ++ +
Sbjct: 417 -------------KNSG----VLDWLMRFRIGLGAARGLAWLHHGCHPPIIQQNICSNVI 545
Query: 703 YLDYDLEPRLSDFGLAKIFGSGLDEEIAR---GSPGYVPPEFSQPEFESPTPKSDVYCFG 759
+D + + R+ DFGLA++ S + G GY+ PE+S S K DVY FG
Sbjct: 546 LVDEEFDARIMDFGLARLMTSDANGSFVNGDLGELGYIAPEYSSTMVAS--LKGDVYGFG 719
Query: 760 VVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEAL 819
V+L EL+TG KP+ + D++ LV WV + ID I G+DE+I + L
Sbjct: 720 VLLLELVTGCKPLEVNNIDEEFKGNLVDWVNMHSSSGRLKDCIDRSISGKGNDEEILQFL 899
Query: 820 KEVGYLCTADLPFKRPTMQQIVGLLKDI 847
K + C R +M Q+ LK I
Sbjct: 900 K-IASNCVIARAKDRWSMYQVYNSLKGI 980
>TC89432 weakly similar to GP|9294592|dbj|BAB02873.1 protein kinase-like
protein {Arabidopsis thaliana}, partial (51%)
Length = 1272
Score = 132 bits (333), Expect(3) = 4e-42
Identities = 79/211 (37%), Positives = 113/211 (53%), Gaps = 2/211 (0%)
Frame = +3
Query: 534 TFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARELEFL 593
T+ +L +AT+ F L EG FG VY G + +AVK L ++ + E A E+E L
Sbjct: 81 TYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEVL 260
Query: 594 GRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADN 653
GR++H NL+ L GYCV DQR+ +YDYM N +L + L+
Sbjct: 261 GRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHG--------------------- 377
Query: 654 GIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLS 713
Q G L W+ R IA+G+A + +LHH +P IIHR +KAS+V LD D P ++
Sbjct: 378 --QYAGEVQL--NWQKRMSIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVA 545
Query: 714 DFGLAKIFGSGLDEEIAR--GSPGYVPPEFS 742
DFG AK+ G+ R G+ GY+ PE++
Sbjct: 546 DFGFAKLIPEGVSHMTTRVKGTLGYLAPEYA 638
Score = 45.4 bits (106), Expect(3) = 4e-42
Identities = 22/65 (33%), Positives = 34/65 (51%)
Frame = +2
Query: 785 LVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKEVGYLCTADLPFKRPTMQQIVGLL 844
+ W L+ K + +DPK+ + Q+++ + V LC P KRP M+Q+V LL
Sbjct: 755 ITEWAEPLITKGRFRDMVDPKLRGNFDENQVKQTVN-VAALCVQSEPEKRPNMKQVVSLL 931
Query: 845 KDIEP 849
K EP
Sbjct: 932 KGQEP 946
Score = 33.1 bits (74), Expect(3) = 4e-42
Identities = 12/19 (63%), Positives = 17/19 (89%)
Frame = +1
Query: 754 DVYCFGVVLFELLTGKKPV 772
DVY FG++L EL+TG+KP+
Sbjct: 667 DVYSFGILLLELVTGRKPI 723
>TC78270 similar to GP|16649103|gb|AAL24403.1 Unknown protein {Arabidopsis
thaliana}, partial (80%)
Length = 1463
Score = 168 bits (426), Expect = 7e-42
Identities = 110/321 (34%), Positives = 159/321 (49%), Gaps = 10/321 (3%)
Frame = +3
Query: 534 TFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARELEFL 593
T+ +L A+ NF + EG FG VY+G L G A+KVL S +E E+ +
Sbjct: 276 TYKELKIASDNFSPANKIGEGGFGSVYKGVLKGGKLAAIKVLSTESKQGVKEFLTEINVI 455
Query: 594 GRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADN 653
IKH NLV L G CV GD RI +Y+Y+EN +L L
Sbjct: 456 SEIKHENLVILYGCCVEGDHRILVYNYLENNSLSQTLL---------------------- 569
Query: 654 GIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLS 713
G + W+ R +I LG AR LAFLH P I+HR +KAS++ LD DL P++S
Sbjct: 570 ---AGGHSNIYFDWQTRRRICLGVARGLAFLHEEVLPHIVHRDIKASNILLDKDLTPKIS 740
Query: 714 DFGLAKIFGSGLDEEIAR--GSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKK- 770
DFGLAK+ S + R G+ GY+ PE++ T K+D+Y FGV+L E+++G+
Sbjct: 741 DFGLAKLIPSYMTHVSTRVAGTIGYLAPEYAIR--GQLTRKADIYSFGVLLVEIVSGRSN 914
Query: 771 -----PVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALK--EVG 823
P+ D Y ++ L + + ++ +D + + EEA K ++
Sbjct: 915 TNTRLPIADQY--------ILETTWQLYERKELAQLVDISL---NGEFDAEEACKILKIA 1061
Query: 824 YLCTADLPFKRPTMQQIVGLL 844
LCT D P RPTM +V +L
Sbjct: 1062LLCTQDTPKLRPTMSSVVKML 1124
>TC80060 similar to PIR|D96574|D96574 hypothetical protein T3F20.24
[imported] - Arabidopsis thaliana, partial (32%)
Length = 1547
Score = 167 bits (424), Expect = 1e-41
Identities = 110/318 (34%), Positives = 158/318 (49%), Gaps = 9/318 (2%)
Frame = +1
Query: 541 ATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARELEFLGRIKHPN 600
AT+NFD + EG FGPVY+G L +AVK L S + E E+ + ++HPN
Sbjct: 181 ATNNFDPKNKIGEGGFGPVYKGVLSDGAVIAVKQLSSKSKQGNREFVNEIGMISALQHPN 360
Query: 601 LVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQNVGS 660
LV L G C+ G+Q + +Y+YMEN +L L+ P
Sbjct: 361 LVKLYGCCIEGNQLLLVYEYMENNSLARALFGKP-------------------------E 465
Query: 661 EGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKI 720
+ L WR R KI +G AR LA+LH I+HR +KA++V LD +L ++SDFGLAK
Sbjct: 466 QRLNLDWRTRMKICVGIARGLAYLHEESRLKIVHRDIKATNVLLDKNLNAKISDFGLAK- 642
Query: 721 FGSGLDEE-------IARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVG 773
LDEE G+ GY+ PE++ + T K+DVY FGVV E+++G
Sbjct: 643 ----LDEEENTHISTRIAGTIGYMAPEYAMRGY--LTDKADVYSFGVVALEIVSGMS--N 798
Query: 774 DDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALK--EVGYLCTADLP 831
+Y +E L+ W L + +DP + GS EEA++ ++ LCT P
Sbjct: 799 TNYRPKEEFVYLLDWAYVLQEQGNLLELVDPTL---GSKYSSEEAMRMLQLALLCTNPSP 969
Query: 832 FKRPTMQQIVGLLKDIEP 849
RP M +V +L+ P
Sbjct: 970 TLRPPMSSVVSMLEGNTP 1023
>TC87606 similar to PIR|C84473|C84473 probable protein kinase [imported] -
Arabidopsis thaliana, partial (69%)
Length = 1803
Score = 164 bits (416), Expect = 1e-40
Identities = 115/330 (34%), Positives = 166/330 (49%), Gaps = 11/330 (3%)
Frame = +3
Query: 530 LLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIH-------VAVKVLVVGSTLT 582
L T A+L T F L G FGPV++GF+ + VAVK+L + S
Sbjct: 444 LYAFTVAELKVITQQFSSSNHLGAGGFGPVHKGFIDDKVRPGLKAQSVAVKLLDLESKQG 623
Query: 583 DEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDD 642
+E E+ LG+++ P+LV L GYC+ + R+ +Y+Y+ G+L+N L+ ++
Sbjct: 624 HKEWLTEVVVLGQLRDPHLVKLIGYCIEDEHRLLVYEYLPRGSLENQLF---RRFSASLP 794
Query: 643 WSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSV 702
WST R KIA+G A+ LAFLH P+I R KAS++
Sbjct: 795 WST------------------------RMKIAVGAAKGLAFLHE-AEQPVIFRDFKASNI 899
Query: 703 YLDYDLEPRLSDFGLAKIFGSGLDEEIA---RGSPGYVPPEFSQPEFESPTPKSDVYCFG 759
LD D +LSDFGLAK G D ++ G+ GY PE+ T KSDVY FG
Sbjct: 900 LLDSDYNAKLSDFGLAKDGPEGDDTHVSTRVMGTQGYAAPEYVMT--GHLTAKSDVYSFG 1073
Query: 760 VVLFELLTGKKPVGDDYTDDKEATTLVSWVRG-LVRKNQTSRAIDPKICDTGSDEQIEEA 818
VVL ELLTG+K V D + LV W R L+ + S+ +DPK+ S+ ++A
Sbjct: 1074VVLLELLTGRKSV--DKNRPQREQNLVDWARPMLIDSRKISKIMDPKLEGQYSEMGAKKA 1247
Query: 819 LKEVGYLCTADLPFKRPTMQQIVGLLKDIE 848
+ Y C + P RPTM +V +L+ ++
Sbjct: 1248-ASLAYQCLSHRPKSRPTMSNVVKILEPLQ 1334
>TC92229 similar to PIR|B86369|B86369 hypothetical protein AAC98010.1
[imported] - Arabidopsis thaliana, partial (44%)
Length = 1347
Score = 164 bits (416), Expect = 1e-40
Identities = 107/317 (33%), Positives = 158/317 (49%), Gaps = 6/317 (1%)
Frame = +2
Query: 534 TFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARELEFL 593
++ +L T+ F ++ EG FG VY+ +P A+K+L GS + E E++ +
Sbjct: 161 SYDQILEITNGFSSENVIGEGGFGRVYKALMPDGRVGALKLLKAGSGQGEREFRAEVDTI 340
Query: 594 GRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADN 653
R+ H +LV L GYC+A QR+ IY+++ NGNL L++ W D
Sbjct: 341 SRVHHRHLVSLIGYCIAEQQRVLIYEFVPNGNLDQHLHE-------------SQWNVLD- 478
Query: 654 GIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLS 713
W R KIA+G AR LA+LH GC+P IIHR +K+S++ LD E +++
Sbjct: 479 -------------WPKRMKIAIGAARGLAYLHEGCNPKIIHRDIKSSNILLDDSYEAQVA 619
Query: 714 DFGLAKIFGSGLDEEIAR--GSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKP 771
DFGLA++ R G+ GY+ PE++ T +SDV+ FGVVL EL+TG+KP
Sbjct: 620 DFGLARLTDDTNTHVSTRVMGTFGYMAPEYATS--GKLTDRSDVFSFGVVLLELVTGRKP 793
Query: 772 VGDDYTDDKEATTLVSWVRGL----VRKNQTSRAIDPKICDTGSDEQIEEALKEVGYLCT 827
V D T +LV W R + + S DP++ D ++ + E C
Sbjct: 794 V--DPTQPVGDESLVEWARPILLRAIETGDFSELADPRLHRQYIDSEMFRMI-EAAAACI 964
Query: 828 ADLPFKRPTMQQIVGLL 844
KRP M QI L
Sbjct: 965 RHSAPKRPRMVQIARAL 1015
>TC83631 similar to PIR|T10659|T10659 probable serine/threonine-specific
protein kinase (EC 2.7.1.-) T5F17.100 - Arabidopsis
thaliana, partial (24%)
Length = 975
Score = 164 bits (415), Expect = 1e-40
Identities = 93/270 (34%), Positives = 149/270 (54%), Gaps = 4/270 (1%)
Frame = +1
Query: 584 EEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDW 643
++ E+ LGR++H N+V L G+ + +Y++M NGNL + ++ G QS
Sbjct: 31 DDLVGEVNLLGRLRHRNIVRLLGFLYNDTDVMIVYEFMVNGNLGDAMH----GKQS---- 186
Query: 644 STDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVY 703
E LL W R+ IALG A+ LA+LHH C PP+IHR +K++++
Sbjct: 187 -----------------ERLLVDWVSRYNIALGIAQGLAYLHHDCHPPVIHRDIKSNNIL 315
Query: 704 LDYDLEPRLSDFGLAKIF-GSGLDEEIARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVL 762
LD +LE R++DFGLAK+ + GS GY+ PE+ K D+Y FG+VL
Sbjct: 316 LDANLEARIADFGLAKMMVRKNETVSMIAGSYGYIAPEYGYS--LKVDEKIDIYSFGIVL 489
Query: 763 FELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEAL--- 819
EL+TGK+P+ D+ E+ +V W+R + KN A+DP + G+ + ++E +
Sbjct: 490 LELITGKRPIDPDF---GESVDIVGWIRRKIDKNSPEEALDPSV---GNCKHVQEEMLLV 651
Query: 820 KEVGYLCTADLPFKRPTMQQIVGLLKDIEP 849
+ LCTA LP +RP+M+ ++ +L + +P
Sbjct: 652 LRIALLCTAKLPKERPSMRDVIMMLGEAKP 741
>TC87497 similar to GP|9759480|dbj|BAB10485.1 serine/threonine protein
kinase-like protein {Arabidopsis thaliana}, partial
(59%)
Length = 1331
Score = 162 bits (409), Expect = 7e-40
Identities = 119/399 (29%), Positives = 189/399 (46%), Gaps = 10/399 (2%)
Frame = +2
Query: 456 LALVLTLSLIFALAGILFLAFGCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADV 515
LA++ ++ F + IL + K + ++ + + + P + T S++ D
Sbjct: 59 LAVLAAIASFFVVTVILAAIILICQHRKTTQTRRNNQIRTRPLRNPNHYPTSSSSLPVDA 238
Query: 516 KQATSVPVVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVL 575
+ P L I+ +L AT NF ++ +G FG VY+ L VAVK L
Sbjct: 239 SWSWD--------PNLKISMEELSRATKNFSNTLIVGDGSFGYVYKASLSTGATVAVKKL 394
Query: 576 VVGSTLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPL 635
+ E A E+E L +++H N+V + GY +G +R+ +Y+++E GNL L++
Sbjct: 395 SPDAFQGFREFAAEMETLSKLRHHNIVKILGYWASGAERLLVYEFIEKGNLDQWLHE--- 565
Query: 636 GVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHR 695
S ST +E I + S W R KI G A L +L HG PIIHR
Sbjct: 566 --SSPTSSSTHQNDEVSISIDFIRSP---LPWETRVKIIRGVAHGLCYL-HGLEKPIIHR 727
Query: 696 AVKASSVYLDYDLEPRLSDFGLAKIFG---SGLDEEIARGSPGYVPPEFSQPEFESPTPK 752
+KAS+V LD + E ++DFGLA+ S + ++A G+ GY+PPE+ PK
Sbjct: 728 DIKASNVLLDSEFEAHIADFGLARRMDKSHSHVSTQVA-GTIGYMPPEYRDGS-NVANPK 901
Query: 753 SDVYCFGVVLFELLTGKKP------VGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKI 806
DVY FGV++ E ++G +P G+D LV+W R + +N +D I
Sbjct: 902 VDVYSFGVLMIETVSGHRPNLAVKLEGND-------IGLVNWARKMKERNTELEMLDGNI 1060
Query: 807 -CDTGSDEQIEEALKEVGYLCTADLPFKRPTMQQIVGLL 844
+ G E+ + +CT +L RP M ++V LL
Sbjct: 1061PREEGLKEESVREYVRIACMCTGELQKDRPEMPEVVKLL 1177
>TC86901 similar to GP|13937147|gb|AAK50067.1 AT5g13160/T19L5_120
{Arabidopsis thaliana}, partial (83%)
Length = 2028
Score = 159 bits (403), Expect = 3e-39
Identities = 111/316 (35%), Positives = 157/316 (49%), Gaps = 5/316 (1%)
Frame = +1
Query: 534 TFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIH-VAVKVLVVGSTLTDEEAARELEF 592
TF +L +AT NF + L EG FG VY+G L VAVK L + E E+
Sbjct: 496 TFRELAAATKNFRPQSFLGEGGFGRVYKGRLETTGQAVAVKQLDRNGLQGNREFLVEVLM 675
Query: 593 LGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEAD 652
L + PNLV L GYC GDQR+ +Y++M G+L++ L+DLP + D
Sbjct: 676 LSLLHSPNLVSLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPADKEPLD----------- 822
Query: 653 NGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRL 712
W R KIA G A+ L +LH +PP+I+R K+S++ LD P+L
Sbjct: 823 --------------WNTRMKIAAGAAKGLEYLHDKANPPVIYRDFKSSNILLDEGYHPKL 960
Query: 713 SDFGLAKIFGSGLDEEIA---RGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGK 769
SDFGLAK+ G ++ G+ GY PE++ T KSDVY FGVV EL+TG+
Sbjct: 961 SDFGLAKLGPVGDKSHVSTRVMGTYGYCAPEYAMT--GQLTVKSDVYSFGVVFLELITGR 1134
Query: 770 KPVGDDYTDDKEATTLVSWVRGLVR-KNQTSRAIDPKICDTGSDEQIEEALKEVGYLCTA 828
K + D T LV+W R L + + S+ DP++ + +AL V +C
Sbjct: 1135KAI--DSTRPHGEQNLVTWARPLFNDRRKFSKLADPRLQGRYPMRGLYQAL-AVASMCIQ 1305
Query: 829 DLPFKRPTMQQIVGLL 844
+ RP + +V L
Sbjct: 1306EQAAARPLIGDVVTAL 1353
>TC77418 similar to GP|15146244|gb|AAK83605.1 At2g17220/T23A1.8 {Arabidopsis
thaliana}, partial (74%)
Length = 1786
Score = 158 bits (399), Expect = 1e-38
Identities = 116/330 (35%), Positives = 164/330 (49%), Gaps = 11/330 (3%)
Frame = +1
Query: 534 TFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIH-----VAVKVLVVGSTLTDEEAAR 588
TFA+L +AT NF LL EG FG VY+G+L + + VAVK L EE
Sbjct: 550 TFAELKTATKNFRLDNLLGEGGFGKVYKGWLESSRNSSGTTVAVKKLNTEGYQGFEEWQS 729
Query: 589 ELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTW 648
E+ FLGR+ HPNLV L GYC + + +Y+YM+ G+L+N L+ VQ
Sbjct: 730 EIHFLGRLYHPNLVKLLGYCYEETELLLVYEYMQRGSLENHLFGRGAAVQP--------- 882
Query: 649 EEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDL 708
W R KIA+G A L+FLH II+R KAS++ LD
Sbjct: 883 ----------------LPWDLRLKIAIGAACGLSFLHTS-DREIIYRDFKASNILLDGSY 1011
Query: 709 EPRLSDFGLAKIFGSGLDEEIA---RGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFEL 765
++SDFGLAK+ S ++ G+PGY PE+ Q KSDVY FGVVL E+
Sbjct: 1012NAKISDFGLAKLGPSASQSHLSTTVMGTPGYAAPEYMQT--GHLYVKSDVYGFGVVLVEI 1185
Query: 766 LTGKKPVGDDYTDDKEATTLVSWVR-GLVRKNQTSRAIDPKICDTGSDEQIEEAL--KEV 822
LTG + V D L W++ L + + + +DP++ G I+ AL ++
Sbjct: 1186LTGLRAV--DLNRPSGRHILTDWIKPELQDRKKLKKVMDPQL---GDKYPIKAALPIAKL 1350
Query: 823 GYLCTADLPFKRPTMQQIVGLLKDIEPTTS 852
C A P RP+M+ ++ L+ I+ T+
Sbjct: 1351AISCLAPEPKLRPSMRDVLERLQGIQAATN 1440
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.137 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,726,439
Number of Sequences: 36976
Number of extensions: 400577
Number of successful extensions: 4928
Number of sequences better than 10.0: 811
Number of HSP's better than 10.0 without gapping: 2884
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3706
length of query: 852
length of database: 9,014,727
effective HSP length: 104
effective length of query: 748
effective length of database: 5,169,223
effective search space: 3866578804
effective search space used: 3866578804
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC122160.2


