

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121245.12 + phase: 0 /pseudo
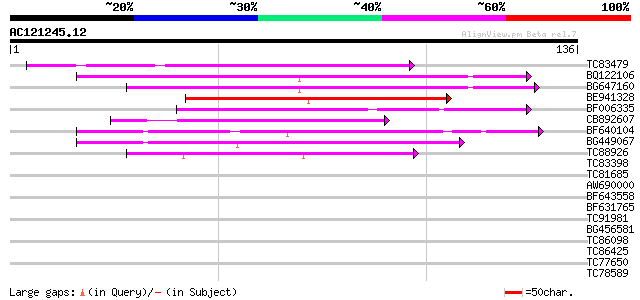
(136 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protei... 68 1e-12
BQ122106 similar to GP|9366656|emb|C probable similar to ring-h2... 67 2e-12
BG647160 homologue to GP|15004435|gb| maturase {Ionopsis satyrio... 51 2e-07
BE941328 48 1e-06
BF006335 similar to GP|14334538|gb| unknown protein {Arabidopsis... 41 2e-04
CB892607 similar to GP|8927657|gb| EST gb|N38213 comes from this... 40 4e-04
BF640104 40 4e-04
BG449067 39 5e-04
TC88926 similar to GP|7290507|gb|AAF45960.1| peb gene product {D... 39 8e-04
TC83398 similar to PIR|T05150|T05150 hypothetical protein F18E5.... 38 0.001
TC81685 38 0.001
AW690000 37 0.002
BF643558 37 0.002
BF631765 33 0.035
TC91981 29 0.50
BG456581 27 2.5
TC86098 similar to GP|14719329|gb|AAK73147.1 putative RING-H2 fi... 27 2.5
TC86425 similar to GP|18854996|gb|AAL79688.1 putative phragmopla... 26 5.6
TC77650 similar to GP|9795587|gb|AAF98405.1| Unknown protein {Ar... 26 5.6
TC78589 1-deoxy-D-xylulose 5-phosphate synthase 2 [Medicago trun... 25 9.5
>TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protein {Plasmodium
falciparum 3D7}, partial (0%)
Length = 1222
Score = 68.2 bits (165), Expect = 1e-12
Identities = 39/93 (41%), Positives = 52/93 (54%)
Frame = +2
Query: 5 LGPTNKGHVICQVRWNPPPEDFIKINVDGSSFGNPDNAGFGGLLRNNRGNWIHGFSGSCG 64
L P ++ + C+ W P + K+N DGS N + AGFGGLLR+ RG I F
Sbjct: 941 LNPVSRSIIWCE--WKKPEIGWTKLNTDGSV--NKETAGFGGLLRDYRGEPICAFVSKAP 1108
Query: 65 RATNLFAELSAIWKGLQLAWDLGYRSIIMESDS 97
+ EL AIW+GL L+ LG +SI +ESDS
Sbjct: 1109 QGDTFLVELWAIWRGLVLSLGLGIKSIWVESDS 1207
>BQ122106 similar to GP|9366656|emb|C probable similar to ring-h2 finger
protein rha1a. {Trypanosoma brucei}, partial (18%)
Length = 693
Score = 67.0 bits (162), Expect = 2e-12
Identities = 42/110 (38%), Positives = 60/110 (54%), Gaps = 1/110 (0%)
Frame = -2
Query: 17 VRWNPPPEDFIKINVDGSSFGNPDNAGFGGLLRNNRGNWIHGFSGSCGRATN-LFAELSA 75
V+WN +NVDGS GNP GF GL+RN G + GF G+ ++ L AEL A
Sbjct: 575 VKWNCDNNSCYILNVDGSCLGNP*PTGFNGLIRNIAGLFNSGFPGNITNTSDILLAELHA 396
Query: 76 IWKGLQLAWDLGYRSIIMESDSQVALDLILDTKQKDFHPHASLLSLIRKL 125
I++GL++ D+G + DS + LI K FH +A+L+ I+ L
Sbjct: 395 IFQGLRMISDMGISDFVCYFDSLHYVSLINGPSMK-FHVYATLIQDIKDL 249
>BG647160 homologue to GP|15004435|gb| maturase {Ionopsis satyrioides},
partial (2%)
Length = 780
Score = 50.8 bits (120), Expect = 2e-07
Identities = 34/100 (34%), Positives = 55/100 (55%), Gaps = 1/100 (1%)
Frame = +1
Query: 29 INVDGSSFGNPDNAGFGGLLRNNRGNWIHGFSGSCGRATN-LFAELSAIWKGLQLAWDLG 87
+NV+ S G P FGG++RN +I GFSG N +FAEL+ + + L+LA L
Sbjct: 331 LNVERSCIGVPIYT*FGGVIRNYLSTYITGFSGFISIYQNIMFAELTTLHQSLKLAISLN 510
Query: 88 YRSIIMESDSQVALDLILDTKQKDFHPHASLLSLIRKLSS 127
++ SDS + ++LI + + H +A L+ I+ + S
Sbjct: 511 IEEMVCYSDSLLTVNLIKEDISQ-HHVYAVLIHNIKYIMS 627
>BE941328
Length = 505
Score = 47.8 bits (112), Expect = 1e-06
Identities = 25/65 (38%), Positives = 43/65 (65%), Gaps = 1/65 (1%)
Frame = +3
Query: 43 GFGGLLRNNRGNWIHGFSGSCGRATNLF-AELSAIWKGLQLAWDLGYRSIIMESDSQVAL 101
G+GG+LRN+ +I FSG +T++ A+L+AI +GL + DL ++ SDS +A+
Sbjct: 3 GYGGILRNSASLFISSFSGFIPNSTDIL*AKLTAIHQGLNMIIDLNMNDVMCYSDSLLAV 182
Query: 102 DLILD 106
+LI++
Sbjct: 183 NLIMN 197
>BF006335 similar to GP|14334538|gb| unknown protein {Arabidopsis thaliana},
partial (3%)
Length = 505
Score = 40.8 bits (94), Expect = 2e-04
Identities = 30/85 (35%), Positives = 42/85 (49%)
Frame = +2
Query: 41 NAGFGGLLRNNRGNWIHGFSGSCGRATNLFAELSAIWKGLQLAWDLGYRSIIMESDSQVA 100
N+GFGGL RN + GF GS G + L AE+ + GL+L W R I DS
Sbjct: 203 NSGFGGLDRNYDKAFQFGFYGSIGWSNILHAEIQDLLVGLKLWWKA--RKFICYYDSLHV 376
Query: 101 LDLILDTKQKDFHPHASLLSLIRKL 125
+ L FH +A++L + + L
Sbjct: 377 VQL-GSMNTHHFHHYANMLEIYQSL 448
>CB892607 similar to GP|8927657|gb| EST gb|N38213 comes from this gene.
{Arabidopsis thaliana}, partial (1%)
Length = 782
Score = 39.7 bits (91), Expect = 4e-04
Identities = 21/67 (31%), Positives = 37/67 (54%)
Frame = +1
Query: 25 DFIKINVDGSSFGNPDNAGFGGLLRNNRGNWIHGFSGSCGRATNLFAELSAIWKGLQLAW 84
D++ +N D NA GGL+++++G+++ ++ G + L AEL I GL + W
Sbjct: 592 DYVVLNCDL-------NAACGGLIQDDQGHFVFHYANKLGSCSVLQAELWGI*HGLSIDW 750
Query: 85 DLGYRSI 91
+ GY I
Sbjct: 751 NRGYSKI 771
>BF640104
Length = 344
Score = 39.7 bits (91), Expect = 4e-04
Identities = 31/115 (26%), Positives = 63/115 (53%), Gaps = 3/115 (2%)
Frame = -1
Query: 17 VRWNPPPEDFIKINVDGSSFGNPDNAGFGGLLRNNRGNWIHGFSGSCGR---ATNLFAEL 73
V+W P + IKIN D ++ + D G G + RN+ G I SG+ R + AE
Sbjct: 344 VKWIKPHQGVIKINCD-ANLTSEDVWGIGVITRNDNG--IVMASGTWNRPGFMCPITAEA 174
Query: 74 SAIWKGLQLAWDLGYRSIIMESDSQVALDLILDTKQKDFHPHASLLSLIRKLSSL 128
+++ A D G+++++ E+D++ + ++ +++++ H + L S+I+ + SL
Sbjct: 173 WGVYQAALFALDQGFQNVLFENDNEKLISML--SREEEGH-RSYLGSIIQSIISL 18
>BG449067
Length = 578
Score = 39.3 bits (90), Expect = 5e-04
Identities = 25/94 (26%), Positives = 47/94 (49%), Gaps = 1/94 (1%)
Frame = -2
Query: 17 VRWNPPPEDFIKINVDGSSFGNPDNAGFGGLLRNNRG-NWIHGFSGSCGRATNLFAELSA 75
V+W P +D IK+N D ++ + D G G + RN+ G G G + AE
Sbjct: 313 VKWKKPEKDIIKLNSD-ANLSSTDLWGIGVVARNDEGFAMASGTWFRFGFPSATTAEAWG 137
Query: 76 IWKGLQLAWDLGYRSIIMESDSQVALDLILDTKQ 109
I++ + A + G+ + ESD++ + ++ T++
Sbjct: 136 IYQAMIFAGEYGFSKVQFESDNERVIQMLNGTEE 35
>TC88926 similar to GP|7290507|gb|AAF45960.1| peb gene product {Drosophila
melanogaster}, partial (1%)
Length = 1073
Score = 38.5 bits (88), Expect = 8e-04
Identities = 24/73 (32%), Positives = 40/73 (53%), Gaps = 3/73 (4%)
Frame = +3
Query: 29 INVDGSSFGNPD--NAGFGGLLRNNRGNWIHGFSGSCGRATNL-FAELSAIWKGLQLAWD 85
+NVDGS + +AG GG+L ++ G W+ GF+ + E AI +GL +
Sbjct: 348 LNVDGSLLREREVPSAGCGGVLSDSSGKWLCGFAQKLNPNLKVDETEKEAILRGLLWVKE 527
Query: 86 LGYRSIIMESDSQ 98
G R I+++SD++
Sbjct: 528 KGKRKILVKSDNE 566
>TC83398 similar to PIR|T05150|T05150 hypothetical protein F18E5.40 -
Arabidopsis thaliana, partial (6%)
Length = 766
Score = 37.7 bits (86), Expect = 0.001
Identities = 28/72 (38%), Positives = 40/72 (54%), Gaps = 1/72 (1%)
Frame = +2
Query: 58 GFSGSCGRATN-LFAELSAIWKGLQLAWDLGYRSIIMESDSQVALDLILDTKQKDFHPHA 116
GFSG + + LFAEL AI GLQLA L ++ SDS ++LI + +H +A
Sbjct: 104 GFSGHIDHSNDILFAELHAILMGLQLAQTLNIVDVVCYSDSLHYVNLI-NGPSVVYHAYA 280
Query: 117 SLLSLIRKLSSL 128
+L+ I+ L L
Sbjct: 281 TLIQDIKDLIRL 316
>TC81685
Length = 1706
Score = 37.7 bits (86), Expect = 0.001
Identities = 20/44 (45%), Positives = 25/44 (56%)
Frame = -2
Query: 17 VRWNPPPEDFIKINVDGSSFGNPDNAGFGGLLRNNRGNWIHGFS 60
V W P D+ KIN DG G+ AG GGL+ + G W+ GFS
Sbjct: 178 VSWTLPQSDWDKINSDGMCQGSL-RAGCGGLI*GDGGEWLCGFS 50
>AW690000
Length = 652
Score = 37.4 bits (85), Expect = 0.002
Identities = 22/64 (34%), Positives = 32/64 (49%)
Frame = +3
Query: 10 KGHVICQVRWNPPPEDFIKINVDGSSFGNPDNAGFGGLLRNNRGNWIHGFSGSCGRATNL 69
K I +V W PP +IK N DGSS ++ GG+ RN+ + + F+ + G
Sbjct: 384 KAPKIIEVIWRPPIPHWIKCNTDGSS--RSHSSACGGIFRNHDTDLLLCFAENTGECNAF 557
Query: 70 FAEL 73
AEL
Sbjct: 558 HAEL 569
>BF643558
Length = 645
Score = 37.0 bits (84), Expect = 0.002
Identities = 17/34 (50%), Positives = 20/34 (58%)
Frame = +2
Query: 10 KGHVICQVRWNPPPEDFIKINVDGSSFGNPDNAG 43
K I +V WNPPP +IK N DGSS N + G
Sbjct: 530 KAPKIIEVLWNPPPLHWIKCNTDGSSNTNTSSCG 631
>BF631765
Length = 498
Score = 33.1 bits (74), Expect = 0.035
Identities = 18/47 (38%), Positives = 24/47 (50%)
Frame = +1
Query: 7 PTNKGHVICQVRWNPPPEDFIKINVDGSSFGNPDNAGFGGLLRNNRG 53
P +G V R PPP IK+NV FG+ + G+G + RN G
Sbjct: 235 PLKRGQVCVVPR*QPPPFGSIKVNVGAGCFGD-GSTGWGFIARNAAG 372
>TC91981
Length = 1224
Score = 29.3 bits (64), Expect = 0.50
Identities = 21/81 (25%), Positives = 34/81 (41%), Gaps = 4/81 (4%)
Frame = -3
Query: 15 CQVRWNPPPEDFIKINVDGSSFGNPDNAGFGGLLRNNRGNWIHGFSGSCGRATNL----F 70
C V W+ P K+N D + + D G ++R+ + + +C L
Sbjct: 568 CNVHWSAPVSGSFKLNADAAGLDDEDRWGLASVVRDAEAVVV---AVACWNRPLLLESDI 398
Query: 71 AELSAIWKGLQLAWDLGYRSI 91
AE A KG + A D+ + SI
Sbjct: 397 AECMATLKG*EFAKDILFLSI 335
>BG456581
Length = 683
Score = 26.9 bits (58), Expect = 2.5
Identities = 11/27 (40%), Positives = 13/27 (47%)
Frame = +2
Query: 17 VRWNPPPEDFIKINVDGSSFGNPDNAG 43
V W PP ++K N DGS N G
Sbjct: 596 VCWKPPSAPWVKGNTDGSXLNNSGAXG 676
>TC86098 similar to GP|14719329|gb|AAK73147.1 putative RING-H2 finger
protein {Oryza sativa}, partial (42%)
Length = 1610
Score = 26.9 bits (58), Expect = 2.5
Identities = 14/38 (36%), Positives = 19/38 (49%)
Frame = -3
Query: 20 NPPPEDFIKINVDGSSFGNPDNAGFGGLLRNNRGNWIH 57
NPPP + +N SS P+ GLLR+ +W H
Sbjct: 831 NPPPGNLHIVNPIASSSI*PETLVSRGLLRSEGSSWRH 718
>TC86425 similar to GP|18854996|gb|AAL79688.1 putative phragmoplastin {Oryza
sativa}, partial (98%)
Length = 2328
Score = 25.8 bits (55), Expect = 5.6
Identities = 11/19 (57%), Positives = 12/19 (62%)
Frame = -3
Query: 96 DSQVALDLILDTKQKDFHP 114
DSQ+A LIL K FHP
Sbjct: 2101 DSQIATKLILPKKPNQFHP 2045
>TC77650 similar to GP|9795587|gb|AAF98405.1| Unknown protein {Arabidopsis
thaliana}, partial (91%)
Length = 1530
Score = 25.8 bits (55), Expect = 5.6
Identities = 17/56 (30%), Positives = 23/56 (40%), Gaps = 5/56 (8%)
Frame = -1
Query: 6 GPTNKGHVICQVRWNPPPED-----FIKINVDGSSFGNPDNAGFGGLLRNNRGNWI 56
GP N HV+ + RW + F + V +FGN G L +R WI
Sbjct: 336 GPINVSHVVNECRWLKTQNERRSFQFFTVLVQTKAFGNHC*LRLG--LNRSRRRWI 175
>TC78589 1-deoxy-D-xylulose 5-phosphate synthase 2 [Medicago truncatula]
Length = 2760
Score = 25.0 bits (53), Expect = 9.5
Identities = 8/12 (66%), Positives = 10/12 (82%)
Frame = -1
Query: 12 HVICQVRWNPPP 23
HVI ++WNPPP
Sbjct: 2130 HVIQNLQWNPPP 2095
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.140 0.453
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,432,472
Number of Sequences: 36976
Number of extensions: 75781
Number of successful extensions: 366
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 360
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 362
length of query: 136
length of database: 9,014,727
effective HSP length: 86
effective length of query: 50
effective length of database: 5,834,791
effective search space: 291739550
effective search space used: 291739550
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 53 (25.0 bits)
Medicago: description of AC121245.12


