

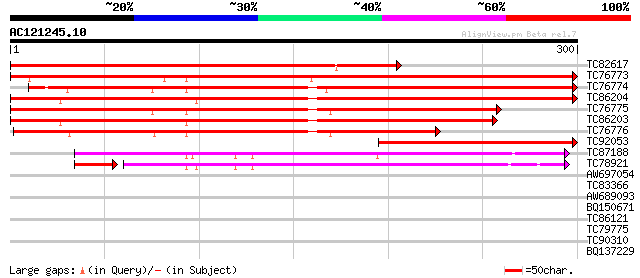
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121245.10 - phase: 0
(300 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC82617 similar to GP|14190477|gb|AAK55719.1 AT5g64260/MSJ1_10 {... 385 e-107
TC76773 similar to GP|3759184|dbj|BAA33810.1 phi-1 {Nicotiana ta... 367 e-102
TC76774 similar to PIR|C85090|C85090 probable phi-1-like phospha... 353 6e-98
TC86204 similar to GP|3759184|dbj|BAA33810.1 phi-1 {Nicotiana ta... 343 4e-95
TC76775 similar to GP|3759184|dbj|BAA33810.1 phi-1 {Nicotiana ta... 298 2e-81
TC86203 similar to PIR|C85090|C85090 probable phi-1-like phospha... 289 8e-79
TC76776 similar to PIR|C85090|C85090 probable phi-1-like phospha... 240 4e-64
TC92053 similar to GP|14190477|gb|AAK55719.1 AT5g64260/MSJ1_10 {... 219 8e-58
TC87188 similar to GP|16604330|gb|AAL24171.1 At2g17230/T23A1.9 {... 174 4e-44
TC78921 similar to GP|16604527|gb|AAL24269.1 AT5g51550/K17N15_10... 153 2e-40
AW697054 similar to GP|9187661|emb| putative class I chitinase {... 28 4.3
TC83366 similar to GP|11994223|dbj|BAB01345. gb|AAF35421.1~gene_... 28 4.3
AW689093 similar to GP|10178248|dbj glucosyltransferase-like pro... 28 5.6
BQ150671 27 7.3
TC86121 similar to GP|15027971|gb|AAK76516.1 unknown protein {Ar... 27 7.3
TC79775 similar to GP|18175886|gb|AAL59945.1 unknown protein {Ar... 27 7.3
TC90310 similar to GP|13937141|gb|AAK50064.1 At1g69210/F4N2_11 {... 27 7.3
BQ137229 similar to GP|21536971|gb| putative chaperonin {Arabido... 27 9.6
>TC82617 similar to GP|14190477|gb|AAK55719.1 AT5g64260/MSJ1_10 {Arabidopsis
thaliana}, partial (47%)
Length = 710
Score = 385 bits (988), Expect = e-107
Identities = 198/210 (94%), Positives = 198/210 (94%), Gaps = 3/210 (1%)
Frame = +1
Query: 1 MASNYYFAIVLSAVLLLVLPTLSFGELVQEQPLVLKYHNGQLLKGKLTVNLIWYGTFTPI 60
MASNYYFAIVLSAVLLLVLPTLSFGELVQEQPLVLKYHNGQLLKGKLTVNLIWYGTFTPI
Sbjct: 82 MASNYYFAIVLSAVLLLVLPTLSFGELVQEQPLVLKYHNGQLLKGKLTVNLIWYGTFTPI 261
Query: 61 QRSIIVDFINSLSTTGAALPSASAWWKTTEKYKVGSSALTVGKQFLHPAYTLGKNLKGKD 120
QRSIIVDFINSLSTTGAALPSASAWWKTTEKYKVGSSALTVGKQFLHPAYTLGKNLKGKD
Sbjct: 262 QRSIIVDFINSLSTTGAALPSASAWWKTTEKYKVGSSALTVGKQFLHPAYTLGKNLKGKD 441
Query: 121 LLALATKFNELSSITVVLTAKDVNVEGFCMSRCGTHGSVRRGSGGARTPYI--WVGNAET 178
LLALATKFNELSSITVVLTAKDVNVEGFCMSRCGTHGSVRRGSGGARTPYI W T
Sbjct: 442 LLALATKFNELSSITVVLTAKDVNVEGFCMSRCGTHGSVRRGSGGARTPYILGW-ETLRT 618
Query: 179 LCPGQCA-WPFHQPIYGPQTPPLVAPNGDV 207
LCPGQCA F PIYGPQTPPLVAPNGDV
Sbjct: 619 LCPGQCAVGLFINPIYGPQTPPLVAPNGDV 708
>TC76773 similar to GP|3759184|dbj|BAA33810.1 phi-1 {Nicotiana tabacum},
partial (86%)
Length = 1228
Score = 367 bits (942), Expect = e-102
Identities = 185/317 (58%), Positives = 227/317 (71%), Gaps = 17/317 (5%)
Frame = +2
Query: 1 MASNYYFAI---VLSAVLLLVLPTLSFGELVQEQPLVLKYHNGQLLKGKLTVNLIWYGTF 57
MAS++ + +L ++ L + ELVQ+Q +L YHNG LL GK++VNLIWYG F
Sbjct: 89 MASSFQSLLKFFLLISIFYLSSAARNLNELVQDQSQLLHYHNGPLLYGKISVNLIWYGNF 268
Query: 58 TPIQRSIIVDFINSLSTTGAALP---SASAWWKTTEKY-------KVGSSALTVGKQFLH 107
P Q++II DF SLS+ ++ P S SAWWKTTEKY K +L++ KQ L
Sbjct: 269 KPSQKAIITDFFTSLSSPSSSKPNQPSVSAWWKTTEKYYHLTSKKKSTQLSLSLNKQILD 448
Query: 108 PAYTLGKNLKGKDLLALATKFNELSSITVVLTAKDVNVEGFCMSRCGTHGS----VRRGS 163
Y+LGK+L K+++ LA+K SI VVLT+ DV+VE FCM RCGTHGS V R
Sbjct: 449 ENYSLGKSLTNKNIIQLASKGEHKDSINVVLTSSDVSVERFCMDRCGTHGSSSSLVPRKG 628
Query: 164 GGARTPYIWVGNAETLCPGQCAWPFHQPIYGPQTPPLVAPNGDVGVDGMVINLATLLAGT 223
A+ YIWVGN+ET CPG CAWPFHQPIYGPQ+ PLVAPN DVG+DGMVINLA+LLAGT
Sbjct: 629 KVAKFAYIWVGNSETQCPGLCAWPFHQPIYGPQSAPLVAPNNDVGLDGMVINLASLLAGT 808
Query: 224 VTNPFNNGYFQGPAAAPLEAVTACTGVFGSGAYPGYAGRVLVDGASGSSYNAHGANGRKY 283
TNPF NGYFQGP+ APLEA +AC GV+G GAYPGYAG +LVD +G+S+NAHG NGRKY
Sbjct: 809 ATNPFGNGYFQGPSEAPLEAASACPGVYGKGAYPGYAGDLLVDSTTGASFNAHGDNGRKY 988
Query: 284 LLPAMWDPQTSACKTLV 300
LLPA++DP T +C TLV
Sbjct: 989 LLPALYDPSTLSCSTLV 1039
>TC76774 similar to PIR|C85090|C85090 probable phi-1-like phosphate-induced
protein [imported] - Arabidopsis thaliana, partial (80%)
Length = 1163
Score = 353 bits (905), Expect = 6e-98
Identities = 186/316 (58%), Positives = 224/316 (70%), Gaps = 26/316 (8%)
Frame = +3
Query: 11 LSAVLLLVLPTLSFGELVQ--------EQPLVLKYHNGQLLKGKLTVNLIWYGTFTPIQR 62
LSA+ L L T+S L+ +Q L +YH G LL GK+++NLIWYG F P QR
Sbjct: 93 LSAIFTLFL-TISLLHLISAARKFTESDQQLKFQYHKGPLLTGKISINLIWYGKFKPSQR 269
Query: 63 SIIVDFINSLST----TGAALPSASAWWKTTEKY-------KVGSSALTVGKQFLHPAYT 111
+II DFI SLS+ T + PS + WWK+TEKY K + AL++G Q L+ Y+
Sbjct: 270 AIITDFITSLSSPKQGTTTSQPSVATWWKSTEKYYQLANNKKSVNLALSLGSQILNENYS 449
Query: 112 LGKNLKGKDLLALATKFNELSSITVVLTAKDVNVEGFCMSRCGTHGSVRRGSGGA----- 166
LGK+L +L LA+K + ++I VVLTA DV V+GFC SRCGTHGS S GA
Sbjct: 450 LGKSLTTNQILKLASKGQQQNAINVVLTAADVLVDGFCSSRCGTHGS----SYGALVNGK 617
Query: 167 --RTPYIWVGNAETLCPGQCAWPFHQPIYGPQTPPLVAPNGDVGVDGMVINLATLLAGTV 224
+ YIWVGN+ET C GQCAWPFHQPIYGPQ+ PLVAPN DVG+DGMVIN+A+LLAGTV
Sbjct: 618 RNKFAYIWVGNSETQCAGQCAWPFHQPIYGPQSAPLVAPNNDVGLDGMVINVASLLAGTV 797
Query: 225 TNPFNNGYFQGPAAAPLEAVTACTGVFGSGAYPGYAGRVLVDGASGSSYNAHGANGRKYL 284
TNPF NGYFQGP APLEA +ACTGV+ GAYPGYAG +LVD SG+SYNA+G NGRKYL
Sbjct: 798 TNPFGNGYFQGPKEAPLEAASACTGVYAKGAYPGYAGDLLVDKTSGASYNANGDNGRKYL 977
Query: 285 LPAMWDPQTSACKTLV 300
LPA+ DP+TSAC TLV
Sbjct: 978 LPAIVDPKTSACSTLV 1025
>TC86204 similar to GP|3759184|dbj|BAA33810.1 phi-1 {Nicotiana tabacum},
partial (76%)
Length = 1242
Score = 343 bits (881), Expect = 4e-95
Identities = 174/308 (56%), Positives = 221/308 (71%), Gaps = 8/308 (2%)
Frame = +3
Query: 1 MASNYYFAIVLSAVLLLVLPTLSFG--ELVQ-EQPLVLKYHNGQLLKGKLTVNLIWYGTF 57
M S+ F+++LS L + L LS +L Q +Q L +YH G LL GK+++NLIWYG F
Sbjct: 93 MTSSTSFSVLLSLFLFISLLHLSSAARKLTQNDQQLKFQYHKGPLLTGKISINLIWYGKF 272
Query: 58 TPIQRSIIVDFINSLST-TGAALPSASAWWKTTEKY-KVGSS---ALTVGKQFLHPAYTL 112
P QR+II DFI S+S+ T PS + WWK T+KY ++ +S LT G L Y+
Sbjct: 273 NPTQRAIISDFIASISSPTIKPQPSVAMWWKLTDKYYQLANSQNLVLTTGSHVLDENYSF 452
Query: 113 GKNLKGKDLLALATKFNELSSITVVLTAKDVNVEGFCMSRCGTHGSVRRGSGGARTPYIW 172
GK+L ++ LA+K ++ ++I VVLT+ DV V+GFC RCG HGS S + Y+W
Sbjct: 453 GKSLTNDQIIKLASKGSQTNAINVVLTSADVVVDGFCSGRCGKHGS----SVNHKFAYVW 620
Query: 173 VGNAETLCPGQCAWPFHQPIYGPQTPPLVAPNGDVGVDGMVINLATLLAGTVTNPFNNGY 232
VGN+ET CPGQCAWPFHQPIYGPQ+PP+VAPN DVG+DGMVIN+A+ LAG +TN F NGY
Sbjct: 621 VGNSETQCPGQCAWPFHQPIYGPQSPPIVAPNNDVGLDGMVINVASWLAGALTNLFGNGY 800
Query: 233 FQGPAAAPLEAVTACTGVFGSGAYPGYAGRVLVDGASGSSYNAHGANGRKYLLPAMWDPQ 292
+QGP APLEA +ACTGV+G GAYPGYAG +LVD SG+SYNA+G NGRKYLLPA++DP
Sbjct: 801 YQGPKEAPLEAASACTGVYGKGAYPGYAGNLLVDPTSGASYNANGINGRKYLLPALFDPT 980
Query: 293 TSACKTLV 300
TS C TLV
Sbjct: 981 TSVCSTLV 1004
>TC76775 similar to GP|3759184|dbj|BAA33810.1 phi-1 {Nicotiana tabacum},
partial (72%)
Length = 926
Score = 298 bits (763), Expect = 2e-81
Identities = 160/279 (57%), Positives = 192/279 (68%), Gaps = 19/279 (6%)
Frame = +2
Query: 1 MASNYYFAIVLSAVLLLVLPTLSFGELVQ-EQPLVLKYHNGQLLKGKLTVNLIWYGTFTP 59
MAS AI + + + +L S +L + +Q L +YH G LL GK++VNLIWYG F P
Sbjct: 53 MASFNKSAIFILFLTISLLQLSSARKLTESDQQLKFQYHKGPLLTGKISVNLIWYGKFKP 232
Query: 60 IQRSIIVDFINSLST----TGAALPSASAWWKTTEKY-------KVGSSALTVGKQFLHP 108
QR+II DFI SLS+ T A+ PS + WWK+TEKY K + AL++G Q L
Sbjct: 233 SQRAIITDFITSLSSPIKPTTASQPSVATWWKSTEKYYQLTNNKKSVNLALSLGSQILDE 412
Query: 109 AYTLGKNLKGKDLLALATKFNELSSITVVLTAKDVNVEGFCMSRCGTHGSVRRGSGGART 168
Y+LGK+L +L LA+K + ++I VVLTA DV V+GFC SRCGTHGS S GAR
Sbjct: 413 KYSLGKSLTTNQILNLASKGQQQNAINVVLTAADVLVDGFCSSRCGTHGS----SYGARV 580
Query: 169 -------PYIWVGNAETLCPGQCAWPFHQPIYGPQTPPLVAPNGDVGVDGMVINLATLLA 221
YIWVGN+ET C GQCAWPFHQPIYGPQ+ PLVAPN DVG+DGMVIN+A LLA
Sbjct: 581 NGKRNKFAYIWVGNSETQCAGQCAWPFHQPIYGPQSAPLVAPNNDVGLDGMVINVARLLA 760
Query: 222 GTVTNPFNNGYFQGPAAAPLEAVTACTGVFGSGAYPGYA 260
GTVTNPF NGYFQGP APLEA +ACTGV+ GAYPGYA
Sbjct: 761 GTVTNPFGNGYFQGPKEAPLEAASACTGVYAKGAYPGYA 877
>TC86203 similar to PIR|C85090|C85090 probable phi-1-like phosphate-induced
protein [imported] - Arabidopsis thaliana, partial (66%)
Length = 862
Score = 289 bits (740), Expect = 8e-79
Identities = 147/266 (55%), Positives = 185/266 (69%), Gaps = 8/266 (3%)
Frame = +3
Query: 1 MASNYYFAIVLSAVLLLVLPTLSFG--ELVQ-EQPLVLKYHNGQLLKGKLTVNLIWYGTF 57
M S+ +++LS L + L LS +L Q +Q L +YH G LL GK+++NL+WYG F
Sbjct: 75 MTSSMSSSVLLSLFLFISLLHLSSAARKLTQNDQQLKFQYHKGPLLTGKISINLVWYGKF 254
Query: 58 TPIQRSIIVDFINSLST-TGAALPSASAWWKTTEKY----KVGSSALTVGKQFLHPAYTL 112
P QR+II DFI S+S+ T PS + WWK T+KY + LT G L Y+
Sbjct: 255 NPSQRAIISDFITSISSPTIKPQPSVATWWKLTDKYYHLANSQNLVLTTGSHILDENYSF 434
Query: 113 GKNLKGKDLLALATKFNELSSITVVLTAKDVNVEGFCMSRCGTHGSVRRGSGGARTPYIW 172
GK+L ++ LA+K ++ ++I VVLT+ DV V+GFC SRCGTHGS S + Y+W
Sbjct: 435 GKSLTNDQIIKLASKGSQTNAINVVLTSADVVVDGFCSSRCGTHGS----SVDHKFAYVW 602
Query: 173 VGNAETLCPGQCAWPFHQPIYGPQTPPLVAPNGDVGVDGMVINLATLLAGTVTNPFNNGY 232
VGN+ET CPGQCAWPFHQPIYGPQ+PPLVAPN DVG+DGMVIN+A+LLAG VTNPF NGY
Sbjct: 603 VGNSETQCPGQCAWPFHQPIYGPQSPPLVAPNNDVGLDGMVINVASLLAGVVTNPFGNGY 782
Query: 233 FQGPAAAPLEAVTACTGVFGSGAYPG 258
+QGP APLEA +ACTG FG GAYPG
Sbjct: 783 YQGPKEAPLEAASACTGEFGKGAYPG 860
>TC76776 similar to PIR|C85090|C85090 probable phi-1-like phosphate-induced
protein [imported] - Arabidopsis thaliana, partial (58%)
Length = 776
Score = 240 bits (613), Expect = 4e-64
Identities = 130/246 (52%), Positives = 163/246 (65%), Gaps = 20/246 (8%)
Frame = +3
Query: 3 SNYYFAIVLSAVLLLVLPTLSFGELVQE--QPLVLKYHNGQLLKGKLTVNLIWYGTFTPI 60
+++ +++ + L + L LS + E Q L ++H G LL GK++VNLIWYG F P
Sbjct: 48 ASFKLSVIFTLFLTISLLQLSSARKLTESDQQLKFQFHKGPLLTGKISVNLIWYGKFKPS 227
Query: 61 QRSIIVDFINSLSTT----GAALPSASAWWKTTEKY-------KVGSSALTVGKQFLHPA 109
QR+II DFI SLS+ PS + WWK+TEKY K + AL++G Q L
Sbjct: 228 QRAIITDFITSLSSPIKPKTTTQPSVATWWKSTEKYYQLTNNKKSVNLALSLGTQILDEK 407
Query: 110 YTLGKNLKGKDLLALATKFNELSSITVVLTAKDVNVEGFCMSRCGTHGSVRRGSGGART- 168
Y+LGK+L +L LA+K + ++I VVLTA DV V+GFC SRCGTHGS S GAR
Sbjct: 408 YSLGKSLTTNQILKLASKGQQQNAINVVLTAADVLVDGFCSSRCGTHGS----SYGARVN 575
Query: 169 ------PYIWVGNAETLCPGQCAWPFHQPIYGPQTPPLVAPNGDVGVDGMVINLATLLAG 222
YIWVGN+ET C GQCAWPFHQPIYGPQ+ PLVAPN DVG+DGMVIN+A+LLAG
Sbjct: 576 GKRNKFAYIWVGNSETQCAGQCAWPFHQPIYGPQSAPLVAPNNDVGLDGMVINVASLLAG 755
Query: 223 TVTNPF 228
TVTNPF
Sbjct: 756 TVTNPF 773
>TC92053 similar to GP|14190477|gb|AAK55719.1 AT5g64260/MSJ1_10 {Arabidopsis
thaliana}, partial (34%)
Length = 571
Score = 219 bits (559), Expect = 8e-58
Identities = 105/105 (100%), Positives = 105/105 (100%)
Frame = +1
Query: 196 QTPPLVAPNGDVGVDGMVINLATLLAGTVTNPFNNGYFQGPAAAPLEAVTACTGVFGSGA 255
QTPPLVAPNGDVGVDGMVINLATLLAGTVTNPFNNGYFQGPAAAPLEAVTACTGVFGSGA
Sbjct: 1 QTPPLVAPNGDVGVDGMVINLATLLAGTVTNPFNNGYFQGPAAAPLEAVTACTGVFGSGA 180
Query: 256 YPGYAGRVLVDGASGSSYNAHGANGRKYLLPAMWDPQTSACKTLV 300
YPGYAGRVLVDGASGSSYNAHGANGRKYLLPAMWDPQTSACKTLV
Sbjct: 181 YPGYAGRVLVDGASGSSYNAHGANGRKYLLPAMWDPQTSACKTLV 315
>TC87188 similar to GP|16604330|gb|AAL24171.1 At2g17230/T23A1.9 {Arabidopsis
thaliana}, partial (82%)
Length = 1285
Score = 174 bits (441), Expect = 4e-44
Identities = 103/278 (37%), Positives = 148/278 (53%), Gaps = 16/278 (5%)
Frame = +3
Query: 35 LKYHNGQLLKGKLTVNLIWYGTFTPIQRSIIVDFINSLSTTGAALPSASAWWKTTEKY-- 92
LKYH G +L + + LIWYG + +S+I DF+NS+S T PS S WW+T Y
Sbjct: 255 LKYHMGPVLSSPINIYLIWYGNWPKPHKSLIKDFLNSISDTTVPHPSVSDWWRTVTLYTD 434
Query: 93 KVG---SSALTVGKQFLHPAYTLGKNLKG---KDLLALATK-----FNELSSITVVLTAK 141
+ G S +++ ++ Y+LGK L +D++A A + + I +VLTA
Sbjct: 435 QTGANISKTVSIAGEYSDYRYSLGKRLTRLSIQDVIATAVRSKPFPVDHRKGIYLVLTAD 614
Query: 142 DVNVEGFCMSRCGTHGSVRRGSGGARTPYIWVGNAETLCPGQCAWPFHQPIY--GPQTPP 199
DV ++ +C + CG H G PY WVGN+ T CP CA+PF P Y G
Sbjct: 615 DVTMDEYCRAVCGFHYFTFPSKVGYTLPYAWVGNSGTQCPEVCAYPFAVPSYMAGGGPGK 794
Query: 200 LVAPNGDVGVDGMVINLATLLAGTVTNPFNNGYFQG-PAAAPLEAVTACTGVFGSGAYPG 258
L PNG+VGVDGMV +A LA +NP N ++ G AP E C G++G+G G
Sbjct: 795 LSPPNGNVGVDGMVSVIAHELAELSSNPLVNAWYAGEDPTAPTEIGDLCEGLYGTGGGGG 974
Query: 259 YAGRVLVDGASGSSYNAHGANGRKYLLPAMWDPQTSAC 296
Y G V+ D SG ++N +G +K+L+ +W +AC
Sbjct: 975 YIGEVMKD-KSGKTFNVNGIRDKKFLVQWVWSNVLNAC 1085
>TC78921 similar to GP|16604527|gb|AAL24269.1 AT5g51550/K17N15_10
{Arabidopsis thaliana}, partial (87%)
Length = 1201
Score = 153 bits (387), Expect(2) = 2e-40
Identities = 92/252 (36%), Positives = 131/252 (51%), Gaps = 16/252 (6%)
Frame = +2
Query: 61 QRSIIVDFINSLSTTGAALPSASAWWKTTEKY--KVGSS---ALTVGKQFLHPAYTLGKN 115
Q+ II +FINS+S PS S WWKT Y + GS+ + +G++ Y+ GK
Sbjct: 197 QKKIIREFINSISAKNTPHPSVSGWWKTVMLYTDQTGSNISNTVHLGQEKNDRFYSHGKT 376
Query: 116 LKG-------KDLLALATK---FNELSSITVVLTAKDVNVEGFCMSRCGTHGSVRRGSGG 165
L K + TK N S + ++LT+ DV V+ FC S CG H G
Sbjct: 377 LTRLSIQSVIKSAIRAKTKPLPINPRSGLYLLLTSDDVYVQDFCTSACGFHYFTFPSLVG 556
Query: 166 ARTPYIWVGNAETLCPGQCAWPFHQPIYGPQTPPLVAPNGDVGVDGMVINLATLLAGTVT 225
PY WVGN+E C GQCA+P+ P + P P +PNGDVGVDGM+ + LA +
Sbjct: 557 YTLPYAWVGNSEKFCAGQCAYPYAVPQFMPNVKPFKSPNGDVGVDGMISVIGHELAELAS 736
Query: 226 NPFNNGYFQ-GPAAAPLEAVTACTGVFGSGAYPGYAGRVLVDGASGSSYNAHGANGRKYL 284
NP N ++ G + P+E C G++G+G Y G+VL D G++YN +G RK+L
Sbjct: 737 NPLANAWYAGGDPSFPVEIADLCEGIYGTGGGGSYTGQVL-DDHDGATYNMNGIR-RKFL 910
Query: 285 LPAMWDPQTSAC 296
+ +W + C
Sbjct: 911 VQWLWSHVLNYC 946
Score = 29.6 bits (65), Expect(2) = 2e-40
Identities = 11/23 (47%), Positives = 16/23 (68%)
Frame = +1
Query: 35 LKYHNGQLLKGKLTVNLIWYGTF 57
L+YH G +L +TV+ IWYG +
Sbjct: 118 LRYHMGPVLTNIITVHTIWYGNW 186
>AW697054 similar to GP|9187661|emb| putative class I chitinase {Phaseolus
vulgaris}, partial (4%)
Length = 815
Score = 28.1 bits (61), Expect = 4.3
Identities = 20/65 (30%), Positives = 25/65 (37%), Gaps = 13/65 (20%)
Frame = -1
Query: 148 FCMSRCGTHG------------SVRRGSGGARTPYIWVGNAETLC-PGQCAWPFHQPIYG 194
F +RCGTH S RR GA + + + PG P H P
Sbjct: 452 FARTRCGTHEACSGDRRVLSSRSRRRDDAGAPLAHTQASRPDPIATPGPQPHPPHPPRPH 273
Query: 195 PQTPP 199
P+TPP
Sbjct: 272 PRTPP 258
>TC83366 similar to GP|11994223|dbj|BAB01345.
gb|AAF35421.1~gene_id:MJK13.21~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (20%)
Length = 650
Score = 28.1 bits (61), Expect = 4.3
Identities = 17/51 (33%), Positives = 29/51 (56%), Gaps = 4/51 (7%)
Frame = +1
Query: 7 FAIVLSAVLLLVLPTLSFGELVQEQP----LVLKYHNGQLLKGKLTVNLIW 53
+ I++ LLL+L TL G LVQE+ + Y+ G+ ++G+ N+ W
Sbjct: 394 YCIIMRIQLLLLLGTLIIGFLVQEEV*LF*AI*GYYKGKFVRGR---NINW 537
>AW689093 similar to GP|10178248|dbj glucosyltransferase-like protein
{Arabidopsis thaliana}, partial (13%)
Length = 471
Score = 27.7 bits (60), Expect = 5.6
Identities = 15/39 (38%), Positives = 21/39 (53%)
Frame = -1
Query: 47 LTVNLIWYGTFTPIQRSIIVDFINSLSTTGAALPSASAW 85
L V+LIWY F P+ + S TTG ++ S+S W
Sbjct: 423 LLVSLIWYVIFIPLLAHLWHSISTSCLTTG-SVESSSTW 310
>BQ150671
Length = 948
Score = 27.3 bits (59), Expect = 7.3
Identities = 16/39 (41%), Positives = 20/39 (51%), Gaps = 2/39 (5%)
Frame = -2
Query: 251 FGSGAYPGYAGRVLVDGASGSSYNA--HGANGRKYLLPA 287
+G AY G V + G + +S HGA G YLLPA
Sbjct: 647 WGRLAYSSLLGAVTLKGEARASMRVPTHGARGSIYLLPA 531
>TC86121 similar to GP|15027971|gb|AAK76516.1 unknown protein {Arabidopsis
thaliana}, partial (89%)
Length = 1882
Score = 27.3 bits (59), Expect = 7.3
Identities = 13/36 (36%), Positives = 17/36 (47%)
Frame = -2
Query: 18 VLPTLSFGELVQEQPLVLKYHNGQLLKGKLTVNLIW 53
VLP L F +L Q PL+ K H L + +W
Sbjct: 993 VLPCLGFQQLSQRTPLICKLHQPHLFHQ*FAESNLW 886
>TC79775 similar to GP|18175886|gb|AAL59945.1 unknown protein {Arabidopsis
thaliana}, partial (82%)
Length = 1147
Score = 27.3 bits (59), Expect = 7.3
Identities = 12/26 (46%), Positives = 17/26 (65%)
Frame = -2
Query: 28 VQEQPLVLKYHNGQLLKGKLTVNLIW 53
V E L++K+HN +LL G T +IW
Sbjct: 702 VGENYLIIKFHNNRLLCGIGTAFIIW 625
>TC90310 similar to GP|13937141|gb|AAK50064.1 At1g69210/F4N2_11 {Arabidopsis
thaliana}, partial (18%)
Length = 723
Score = 27.3 bits (59), Expect = 7.3
Identities = 15/34 (44%), Positives = 20/34 (58%)
Frame = +3
Query: 10 VLSAVLLLVLPTLSFGELVQEQPLVLKYHNGQLL 43
+ S VLLL LPTLSF ++V +L + LL
Sbjct: 138 LFSYVLLLALPTLSFSKIVISLHQILTFSLSPLL 239
>BQ137229 similar to GP|21536971|gb| putative chaperonin {Arabidopsis
thaliana}, partial (10%)
Length = 976
Score = 26.9 bits (58), Expect = 9.6
Identities = 9/18 (50%), Positives = 12/18 (66%)
Frame = +2
Query: 79 LPSASAWWKTTEKYKVGS 96
LPS S WWK +Y++ S
Sbjct: 215 LPSGSPWWKELARYRMSS 268
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.136 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,013,249
Number of Sequences: 36976
Number of extensions: 147758
Number of successful extensions: 724
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 692
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 698
length of query: 300
length of database: 9,014,727
effective HSP length: 96
effective length of query: 204
effective length of database: 5,465,031
effective search space: 1114866324
effective search space used: 1114866324
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC121245.10


