

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121241.13 - phase: 0 /pseudo
(869 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
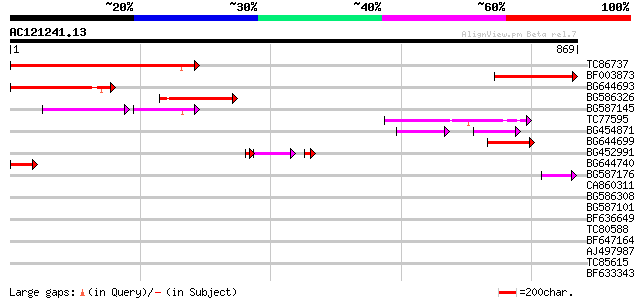
Score E
Sequences producing significant alignments: (bits) Value
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 244 1e-64
BF003873 similar to GP|14715222|em putative polyprotein {Cicer a... 210 2e-54
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 165 6e-41
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 157 2e-38
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 93 6e-33
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 94 3e-19
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 68 5e-15
BG644699 similar to PIR|T07863|T078 probable polyprotein - pinea... 64 2e-10
BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, pa... 50 3e-08
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 52 1e-06
BG587176 weakly similar to PIR|G84493|G84 probable retroelement ... 46 7e-05
CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product ... 40 0.003
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 35 0.094
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 33 0.47
BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativ... 33 0.47
TC80588 similar to GP|23617092|dbj|BAC20775. putative ubiquitin ... 32 1.4
BF647164 weakly similar to GP|6691191|gb F7F22.15 {Arabidopsis t... 30 4.0
AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis... 30 4.0
TC85615 similar to GP|10178248|dbj|BAB11680. glucosyltransferase... 30 5.2
BF633343 29 6.8
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 244 bits (622), Expect = 1e-64
Identities = 127/296 (42%), Positives = 185/296 (61%), Gaps = 7/296 (2%)
Frame = +1
Query: 2 IDYRQLNKVTIKNRYPLPRIDDLMDQLVGARVFSKIDLSSGYHQIKVKDEDMQKTAFRTR 61
+DYR LN +T K+RYPLP I + + ++ GAR F+K+D+ + +H++++KDED +KTAFRTR
Sbjct: 625 VDYRALNAITKKDRYPLPLISETLRRVAGARWFTKLDVVAAFHKMRIKDEDQEKTAFRTR 804
Query: 62 YGHYEYKVMPFGVTNAPGVFMEYMNRIFHAYLDKFVVVFSDDILIYSK-NEEEHAEHMKI 120
YG +E+ V PFG+T AP F Y+N+ H +LD FV + DD+LIY+ ++++H ++
Sbjct: 805 YGLFEWIVCPFGLTGAPATFQRYINKTLHEFLDDFVTAYIDDVLIYTTGSKKDHEAQVRR 984
Query: 121 VLQLLKEKKLYAKLSKCEFWLSEVSFLGHII-SGSGIVVDPSKVDAVSQWETPKSVTEIR 179
VL+ L + L KCEF ++ V ++G I+ +G G+ DP K+ A+ W P SV R
Sbjct: 985 VLRRLADAGLSLDPKKCEFSVTTVKYVGFILTAGKGVSCDPLKLAAIRDWLPPGSVKGAR 1164
Query: 180 SFLGLAGYYRRFIEGFSKLALPLTQLTCKGKSFVWDAQCENSFNELKRRLTTAPILILPK 239
SFLG YY+ FI G+S++ PLT+LT K F W A+ E +F +LKR P+L +
Sbjct: 1165SFLGFCNYYKDFIPGYSEITEPLTRLTRKDFPFRWGAEQEAAFTKLKRLFAEEPVLRMFD 1344
Query: 240 PDEPFVVYCDASKLGLGGVLMQD-----GKVVAYASRQLRIHEKNYPTHDLELAAV 290
P+ V D S LGGVL Q+ VA+ S++L E NYP HD EL AV
Sbjct: 1345PEAVTTVETDCSGFALGGVLTQEDGTGAAHPVAFHSQRLSPAEYNYPIHDKELLAV 1512
>BF003873 similar to GP|14715222|em putative polyprotein {Cicer arietinum},
partial (82%)
Length = 559
Score = 210 bits (535), Expect = 2e-54
Identities = 105/126 (83%), Positives = 113/126 (89%)
Frame = +2
Query: 744 GVGRALKSKKLTPKFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADPSHV 803
GVGRALKSKKLT +FIGPYQISERVGTVAYRVGLPPHL NLHDVFHVSQLRKYV DPSHV
Sbjct: 2 GVGRALKSKKLTVRFIGPYQISERVGTVAYRVGLPPHLLNLHDVFHVSQLRKYVPDPSHV 181
Query: 804 IPRADVQVRDNLTIETLPLRIEYREVKKLRGKDIPLVKVVLGGVTGESLTWELESKMLES 863
I DVQVRDNLT+ETLP+RI+ R+VK LRGK+IPLV+VV GESLTWELESKM+ES
Sbjct: 182 IQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWDRANGESLTWELESKMVES 361
Query: 864 YPELFA 869
YPELFA
Sbjct: 362 YPELFA 379
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 165 bits (418), Expect = 6e-41
Identities = 89/167 (53%), Positives = 110/167 (65%), Gaps = 5/167 (2%)
Frame = +2
Query: 1 TIDYRQLNKVTIKNRYPLPRIDDLMDQLVGARVFSKIDLSSGYHQIKVKDEDMQKTAFRT 60
+IDY QLN V IK +YPLP ID+L D L G++ F KIDL G HQ +V ED+ KTAFR
Sbjct: 230 SIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVIGEDVPKTAFRI 409
Query: 61 RYGHYEYKVMPFGVTNAPGVFMEYMNRIFHAYLDKFVVVFSDDILIYSKNEEEHAEHMKI 120
RYGHYE VM FG TN P FME MNR+F YLD V+VFS+DILIYSKNE EH H+++
Sbjct: 410 RYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDILIYSKNENEHENHLRL 589
Query: 121 VLQLLKEKKLYAKLSKCEF----WLSEVSFLG-HIISGSGIVVDPSK 162
L++LK+ + C+ L EV F H+ISG G+ VD +
Sbjct: 590 ALKVLKD------IGLCQISYV*ILVEVGFFSLHVISGEGLKVDSKR 712
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 157 bits (396), Expect = 2e-38
Identities = 77/119 (64%), Positives = 93/119 (77%)
Frame = +2
Query: 230 TTAPILILPKPDEPFVVYCDASKLGLGGVLMQDGKVVAYASRQLRIHEKNYPTHDLELAA 289
T+APIL+LP+ +VVY DAS GLG VL Q KV+AYASRQLR HE NYPTHDLE+AA
Sbjct: 8 TSAPILVLPELIT-YVVYTDASITGLGCVLTQHEKVIAYASRQLRKHEGNYPTHDLEMAA 184
Query: 290 VVFVLKIWRHYLYGS*FEVFSDHKSLKYLFDQKELNMRQRRWLELLKDCDFGLNYHPGK 348
VVF LKIWR YLYG+ ++ +DHKSLKY+F Q ELN+RQRRW+E + D D + Y+PGK
Sbjct: 185 VVFALKIWRSYLYGAKVQIHTDHKSLKYIFTQPELNLRQRRWMEFVADYDLDITYYPGK 361
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 92.8 bits (229), Expect(2) = 6e-33
Identities = 47/133 (35%), Positives = 76/133 (56%)
Frame = +2
Query: 51 EDMQKTAFRTRYGHYEYKVMPFGVTNAPGVFMEYMNRIFHAYLDKFVVVFSDDILIYSKN 110
+D++KTAF T G Y YKVMPFG+ NA + +NR+F L + V+ DD+L+ S
Sbjct: 11 DDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVKSLR 190
Query: 111 EEEHAEHMKIVLQLLKEKKLYAKLSKCEFWLSEVSFLGHIISGSGIVVDPSKVDAVSQWE 170
+H H+K + L E + +KC F ++ FLG+I++ GI V+P ++ A+
Sbjct: 191 ATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAILDLP 370
Query: 171 TPKSVTEIRSFLG 183
+PK+ E++ G
Sbjct: 371 SPKNSREVQRLTG 409
Score = 67.4 bits (163), Expect(2) = 6e-33
Identities = 36/106 (33%), Positives = 56/106 (51%), Gaps = 4/106 (3%)
Frame = +3
Query: 190 RFIEGFSKLALPLTQLTCKGKSFVWDAQCENSFNELKRRLTTAPILILPKPDEPFVVYCD 249
RFI + LP +L C K FVWD +CE +F +LK+ LTT P+L P+ + +Y
Sbjct: 429 RFISRSTDKCLPFYKLLCGNKRFVWDEKCEEAFEQLKQYLTTPPVLSKPEAGDTLSLYIA 608
Query: 250 ASKLGLGGVLMQDG----KVVAYASRQLRIHEKNYPTHDLELAAVV 291
S + VL+++ K + Y S+++ E YPT + AV+
Sbjct: 609 ISSTAVSSVLIREDRGEQKPIFYTSKRMTDPETRYPTLEKMAFAVI 746
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial
(14%)
Length = 1708
Score = 93.6 bits (231), Expect = 3e-19
Identities = 68/232 (29%), Positives = 109/232 (46%), Gaps = 6/232 (2%)
Frame = +2
Query: 575 KLHGVPSSIVSDRNFRFTSRFWKSLQDALGSKLRVSSAYHPQADGHSERTIQSLEDLLRV 634
+ HG+P SIVSDR + RFW+ G +S++YHPQ DG +ER Q ++ +LR
Sbjct: 554 RFHGMPQSIVSDRGSNWVGRFWREFCRLTGVTQLLSTSYHPQTDGGTERWNQEIQAVLRA 733
Query: 635 CVLEQGGAWDSHLPLIEFTYNNSYHSSIGMAPFETLYGRRCRTPLCWFESGESVLLGPDL 694
V W LP ++ N ++SSIG PF +G P+ E V+ +
Sbjct: 734 YVCWSQDNWGDLLPTVQLALRNRHNSSIGATPFFVEHGYHV-DPIPTVEDTGGVVSEGEA 910
Query: 695 VHQTIEK-----VQMIREKMKASQSRQKSYHDKRK-KALEFQEGDHVFLRVTPMTGVGRA 748
Q + K I+ ++ A+Q R ++ +KR+ A +Q GD V+L V+ +
Sbjct: 911 AAQLLVKRMKDVTGFIQAEIVAAQQRSEASANKRRCPADRYQVGDKVWLNVSNYKSPRPS 1090
Query: 749 LKSKKLTPKFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADP 800
K L K Y+++ V + +P ++ FHV LR+ +DP
Sbjct: 1091KKLDWLHHK----YEVTRFVTPHVVELNVP---GTVYPKFHVDLLRRAASDP 1225
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 67.8 bits (164), Expect(2) = 5e-15
Identities = 35/81 (43%), Positives = 43/81 (52%)
Frame = +2
Query: 593 SRFWKSLQDALGSKLRVSSAYHPQADGHSERTIQSLEDLLRVCVLEQGGAWDSHLPLIEF 652
S FWK L G+ L +SSAYHP +DG SE + E LR + W P E+
Sbjct: 32 SNFWKQLFKLHGTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEY 211
Query: 653 TYNNSYHSSIGMAPFETLYGR 673
YN SY+ S M PF+ LYGR
Sbjct: 212 WYNTSYNISAAMTPFKALYGR 274
Score = 32.0 bits (71), Expect(2) = 5e-15
Identities = 21/73 (28%), Positives = 34/73 (45%), Gaps = 2/73 (2%)
Frame = +1
Query: 712 SQSRQKSYHDKRKKALEFQEGDHVFLRVTPMTGVGRALK--SKKLTPKFIGPYQISERVG 769
+Q K DK+++ EFQ G+HV +++ P AL+ K +P F +
Sbjct: 388 AQQTMKHQADKKRRHFEFQLGEHVLVKLQPYQQSSVALRKYQKFGSPNFGSLLTVCSL*V 567
Query: 770 TVAYRVGLPPHLS 782
A+ PP+LS
Sbjct: 568 ESAFHCKSPPYLS 606
>BG644699 similar to PIR|T07863|T078 probable polyprotein - pineapple
retrotransposon dea1 (fragment), partial (5%)
Length = 231
Score = 63.9 bits (154), Expect = 2e-10
Identities = 31/73 (42%), Positives = 48/73 (65%), Gaps = 1/73 (1%)
Frame = +2
Query: 733 DHVFLRVTPMT-GVGRALKSKKLTPKFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVS 791
+ V L+V P G R K KL+ ++IGP+++ +R+G VAY + LPP LS +H VFHVS
Sbjct: 2 EQVLLKVLPTERGDCRFGKRGKLSLRYIGPFEVIKRIGEVAYELALPPGLSGVHPVFHVS 181
Query: 792 QLRKYVADPSHVI 804
++Y D +++I
Sbjct: 182 MFKRYHGDGNYII 220
>BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, partial (33%)
Length = 560
Score = 50.4 bits (119), Expect(3) = 3e-08
Identities = 34/65 (52%), Positives = 39/65 (59%)
Frame = +1
Query: 374 LNSSET*V*FVSCHLRVCNWVC*RLIVIS*TIS*KRRKLMSSLWI*WLLVIELKRVTSRL 433
LNSSET*V FV C RV NW C*R +S + R+ M S I* L +I LK V +L
Sbjct: 70 LNSSET*VWFVKCRHRV*NWGC*RSTTNFWIVSKRLRRWM*SW*I*CLGIIRLKMVILKL 249
Query: 434 MIKVC 438
MIK C
Sbjct: 250 MIKEC 264
Score = 23.9 bits (50), Expect(3) = 3e-08
Identities = 10/13 (76%), Positives = 11/13 (83%)
Frame = +3
Query: 362 TCLL*W*KSLSCL 374
TCLL*W +S SCL
Sbjct: 33 TCLL*WLESWSCL 71
Score = 21.6 bits (44), Expect(3) = 3e-08
Identities = 12/16 (75%), Positives = 13/16 (81%)
Frame = +3
Query: 453 *KS*F*KKVTEVA*VF 468
*K *F*KKV EV *+F
Sbjct: 285 *KR*F*KKVIEVM*IF 332
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 51.6 bits (122), Expect = 1e-06
Identities = 25/41 (60%), Positives = 29/41 (69%)
Frame = -1
Query: 2 IDYRQLNKVTIKNRYPLPRIDDLMDQLVGARVFSKIDLSSG 42
IDYRQ NKVT KN+YPLPRID+L D++ F IDL G
Sbjct: 190 IDYRQFNKVTTKNKYPLPRIDNLFDKIQEDCYF*NIDLRLG 68
>BG587176 weakly similar to PIR|G84493|G84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(1%)
Length = 729
Score = 45.8 bits (107), Expect = 7e-05
Identities = 24/54 (44%), Positives = 32/54 (58%)
Frame = -1
Query: 815 LTIETLPLRIEYREVKKLRGKDIPLVKVVLGGVTGESLTWELESKMLESYPELF 868
L +ET P+RI R K +R K I +VK+V E +TWE E++M YPE F
Sbjct: 717 LDLETRPVRILDRMEKAMRKKPIQMVKIVWDCSGREEITWETEARMKADYPEWF 556
>CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product {Drosophila
melanogaster}, partial (20%)
Length = 192
Score = 40.4 bits (93), Expect = 0.003
Identities = 18/43 (41%), Positives = 28/43 (64%)
Frame = +1
Query: 266 VAYASRQLRIHEKNYPTHDLELAAVVFVLKIWRHYLYGS*FEV 308
+AYASR L E+NY + E A ++ ++ +RHYL+G FE+
Sbjct: 64 IAYASRLLTAAERNYTVVERECLAAIWAIRNFRHYLHGPKFEL 192
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 686
Score = 35.4 bits (80), Expect = 0.094
Identities = 19/71 (26%), Positives = 36/71 (49%)
Frame = -2
Query: 577 HGVPSSIVSDRNFRFTSRFWKSLQDALGSKLRVSSAYHPQADGHSERTIQSLEDLLRVCV 636
HG+P IV+D F S ++ + +L +S +PQ++G +E + + + D L+ +
Sbjct: 685 HGLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKIIIDGLKKRL 506
Query: 637 LEQGGAWDSHL 647
+ G W L
Sbjct: 505 DLKKGCWADEL 473
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 33.1 bits (74), Expect = 0.47
Identities = 14/39 (35%), Positives = 24/39 (60%)
Frame = +2
Query: 578 GVPSSIVSDRNFRFTSRFWKSLQDALGSKLRVSSAYHPQ 616
GVP ++SD F ++ ++ L G + +V++AYHPQ
Sbjct: 482 GVPRVVISDGGSHFINKVFEKLLKKNGVRHKVATAYHPQ 598
>BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativa}, partial
(4%)
Length = 653
Score = 33.1 bits (74), Expect = 0.47
Identities = 34/156 (21%), Positives = 62/156 (38%), Gaps = 4/156 (2%)
Frame = +1
Query: 627 SLEDLLRVCVLEQGGAWDSHLPLIEFTYNNSYHSSIGMAPFETLYGRRCRTPLCWFESGE 686
+LE L EQ G + L E YN ++H + G PF+ +Y L F
Sbjct: 37 TLETHLLYFTSEQQGV*NFFLTWAECLYNTNFHRTAGCTPFKVVY---VVAHLQKFVVAR 207
Query: 687 SVLLGPDLVHQTIEKVQMIREKMKASQSRQKSY----HDKRKKALEFQEGDHVFLRVTPM 742
++ + +H++ R + +Y H +R ++ + +V P
Sbjct: 208 DLIYRNEGLHKSST*TSFGRGTRAYEALSRPAYETC*HPRRPLSIVYTRDRTYEWQVLP- 384
Query: 743 TGVGRALKSKKLTPKFIGPYQISERVGTVAYRVGLP 778
K GPYQ+ +++G+VA+++ LP
Sbjct: 385 ----------KYVA*CYGPYQVIKQIGSVAFKL*LP 462
>TC80588 similar to GP|23617092|dbj|BAC20775. putative ubiquitin
carboxyl-terminal hydrolase {Oryza sativa (japonica
cultivar-group)}, partial (30%)
Length = 1110
Score = 31.6 bits (70), Expect = 1.4
Identities = 20/66 (30%), Positives = 35/66 (52%)
Frame = +2
Query: 73 GVTNAPGVFMEYMNRIFHAYLDKFVVVFSDDILIYSKNEEEHAEHMKIVLQLLKEKKLYA 132
G N P F +Y N AY+ ++ D +I + +E++ AEH++ L+ +E+K +
Sbjct: 341 GFNNPPFKFTKYSN----AYMLVYIRESDKDKIICNVDEKDIAEHLRERLKKEQEEKEHK 508
Query: 133 KLSKCE 138
K K E
Sbjct: 509 KKEKAE 526
>BF647164 weakly similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana},
partial (6%)
Length = 469
Score = 30.0 bits (66), Expect = 4.0
Identities = 21/69 (30%), Positives = 33/69 (47%)
Frame = +1
Query: 707 EKMKASQSRQKSYHDKRKKALEFQEGDHVFLRVTPMTGVGRALKSKKLTPKFIGPYQISE 766
E K + R K +HD+R EF+EG+ V L + + L KL + GP+Q+
Sbjct: 142 ENAKIYKERTKKWHDRRIIRREFREGELVLLFNSRL-----KLFPGKLRSHWSGPFQVKN 306
Query: 767 RVGTVAYRV 775
+ + A V
Sbjct: 307 VMPSGAVEV 333
>AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 636
Score = 30.0 bits (66), Expect = 4.0
Identities = 16/51 (31%), Positives = 25/51 (48%)
Frame = -2
Query: 289 AVVFVLKIWRHYLYGS*FEVFSDHKSLKYLFDQKELNMRQRRWLELLKDCD 339
A+ + K RHY+ + S +KY+F++ L R RW LL + D
Sbjct: 623 ALAWAAKRLRHYMINHTTWLVSKMDPIKYIFEKPALTGRIARWQMLLSEYD 471
>TC85615 similar to GP|10178248|dbj|BAB11680. glucosyltransferase-like protein
{Arabidopsis thaliana}, partial (90%)
Length = 2067
Score = 29.6 bits (65), Expect = 5.2
Identities = 20/54 (37%), Positives = 28/54 (51%)
Frame = -3
Query: 97 VVVFSDDILIYSKNEEEHAEHMKIVLQLLKEKKLYAKLSKCEFWLSEVSFLGHI 150
VVVFS +I+I S EE+H + L+E K + K C ++ FLG I
Sbjct: 1759 VVVFSMNIVIASTKEEKHCK------S*LQEIKPFHKFGSCFLLNFDIGFLGCI 1616
>BF633343
Length = 498
Score = 29.3 bits (64), Expect = 6.8
Identities = 16/36 (44%), Positives = 20/36 (55%)
Frame = +1
Query: 153 GSGIVVDPSKVDAVSQWETPKSVTEIRSFLGLAGYY 188
GSG+++ P +DA S WE SFLG AG Y
Sbjct: 199 GSGLLISPPLLDARSCWED--------SFLGSAGSY 282
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.337 0.148 0.477
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,569,338
Number of Sequences: 36976
Number of extensions: 433875
Number of successful extensions: 2987
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 1599
Number of HSP's successfully gapped in prelim test: 146
Number of HSP's that attempted gapping in prelim test: 1354
Number of HSP's gapped (non-prelim): 1802
length of query: 869
length of database: 9,014,727
effective HSP length: 105
effective length of query: 764
effective length of database: 5,132,247
effective search space: 3921036708
effective search space used: 3921036708
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC121241.13


