

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121241.11 + phase: 0 /pseudo
(175 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
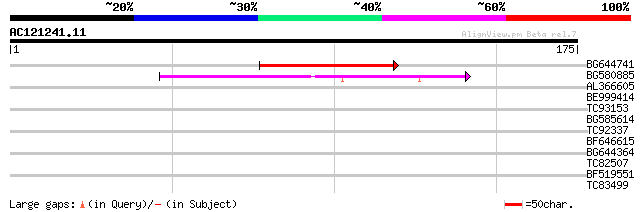
Score E
Sequences producing significant alignments: (bits) Value
BG644741 46 9e-06
BG580885 43 6e-05
AL366605 38 0.002
BE999414 35 0.020
TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Ci... 27 3.2
BG585614 similar to GP|22295990|dbj ORF_ID:tlr2263~hypothetical ... 27 3.2
TC92337 homologue to GP|8572250|gb|AAF77064.1| beta 1 2-xylosylt... 27 4.1
BF646615 homologue to PIR|T48395|T483 GT2-like protein - Arabido... 27 5.4
BG644364 similar to PIR|E86221|E86 hypothetical protein [importe... 27 5.4
TC82507 26 7.1
BF519551 26 9.2
TC83499 26 9.2
>BG644741
Length = 735
Score = 45.8 bits (107), Expect = 9e-06
Identities = 19/43 (44%), Positives = 27/43 (62%)
Frame = -2
Query: 78 LHLFPFSLRDRASAWFHSLKIGSIISWDQMRKAFLSQFFPHSK 120
L +FP SL A WF L SI +W+Q+R FL++++P SK
Sbjct: 587 LRVFPLSLMGEADIWFTELPYNSIFTWNQLRDVFLARYYPVSK 459
>BG580885
Length = 609
Score = 43.1 bits (100), Expect = 6e-05
Identities = 26/102 (25%), Positives = 51/102 (49%), Gaps = 6/102 (5%)
Frame = +3
Query: 47 FFGSPTEDPNLHVSIFLRLSGTLKANQEAVRLHLFPFSLRDRASAWFHSLKIGSI---IS 103
F G+P E P H+S F ++ A+ ++ +FP +L + ++ W+ L I +S
Sbjct: 54 FRGTPNESPITHLSRFNKVCRANNASSVEMQKKIFPVTLEEESALWY-DLNIEPYYISLS 230
Query: 104 WDQMRKAFLSQFFPHSKTAQLR---IKSHSSLKKMVNLFTMR 142
WD+++ +FL ++ +LR + H K+ V + +R
Sbjct: 231 WDEIKLSFLQAYYEIEPVEELRSELMGIHQGEKERVRSYFLR 356
>AL366605
Length = 422
Score = 37.7 bits (86), Expect = 0.002
Identities = 19/61 (31%), Positives = 34/61 (55%)
Frame = -3
Query: 55 PNLHVSIFLRLSGTLKANQEAVRLHLFPFSLRDRASAWFHSLKIGSIISWDQMRKAFLSQ 114
P H+ ++R G K N +++ +H F SL + A+ W+ SL I ++D++ AF S
Sbjct: 291 PQNHIIKYVRKMGNYKDN-DSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDELAAAFKSH 115
Query: 115 F 115
+
Sbjct: 114 Y 112
>BE999414
Length = 613
Score = 34.7 bits (78), Expect = 0.020
Identities = 24/89 (26%), Positives = 44/89 (48%)
Frame = +3
Query: 52 TEDPNLHVSIFLRLSGTLKANQEAVRLHLFPFSLRDRASAWFHSLKIGSIISWDQMRKAF 111
T DP+ H+ + + T ++ + AV+ LF +LR A WF +L+ SI SW + F
Sbjct: 27 T*DPDEHMEN-IEVVLTYRSVRGAVKCKLFVTTLRRGAMTWFKNLRRNSIGSWGDLCHEF 203
Query: 112 LSQFFPHSKTAQLRIKSHSSLKKMVNLFT 140
+ F ++ + S ++ ++N T
Sbjct: 204 TTHFTVSRTQPKMDLFS*LNMSLILNFVT 290
>TC93153 similar to GP|14715220|emb|CAC44106. gag polyprotein {Cicer
arietinum}, partial (8%)
Length = 516
Score = 27.3 bits (59), Expect = 3.2
Identities = 13/48 (27%), Positives = 24/48 (49%), Gaps = 5/48 (10%)
Frame = +2
Query: 85 LRDRASAWFHSL-----KIGSIISWDQMRKAFLSQFFPHSKTAQLRIK 127
L + A W+ SL + ++++W RK FL ++FP + I+
Sbjct: 254 LAEEADDWWISLLPVLEQDDAVVTWAMFRKEFLGRYFPEDVRGKKEIE 397
>BG585614 similar to GP|22295990|dbj ORF_ID:tlr2263~hypothetical protein
{Thermosynechococcus elongatus BP-1}, partial (5%)
Length = 617
Score = 27.3 bits (59), Expect = 3.2
Identities = 13/33 (39%), Positives = 19/33 (57%)
Frame = +1
Query: 12 IISSDEPYDAILYPTVEGNNFEIKSALIYLVQE 44
++SS EP D + V NNF +K IYL+ +
Sbjct: 31 LVSSIEPGDKMEVVVVFENNFIVKKTAIYLIYD 129
>TC92337 homologue to GP|8572250|gb|AAF77064.1| beta 1 2-xylosyltransferase
{Arabidopsis thaliana}, partial (4%)
Length = 448
Score = 26.9 bits (58), Expect = 4.1
Identities = 17/62 (27%), Positives = 27/62 (43%), Gaps = 1/62 (1%)
Frame = +1
Query: 32 FEIKSALIYLVQENQFFGSPTEDPNLHVSIFLRLSGTLKANQEAVRLHLF-PFSLRDRAS 90
F S +Y + F +PT PN H+ IF + S + E + H+ P+ + S
Sbjct: 91 FNSFSLCLYFIHHPSFL-NPTPSPNSHIKIFTKDSPFRRPFIEKHKTHILKPWPILPSYS 267
Query: 91 AW 92
W
Sbjct: 268 PW 273
>BF646615 homologue to PIR|T48395|T483 GT2-like protein - Arabidopsis
thaliana, partial (16%)
Length = 654
Score = 26.6 bits (57), Expect = 5.4
Identities = 15/37 (40%), Positives = 22/37 (58%)
Frame = -1
Query: 108 RKAFLSQFFPHSKTAQLRIKSHSSLKKMVNLFTMRGS 144
++ FLS FF +LRIK+ S+LKK +L + S
Sbjct: 435 KRTFLSSFF------ELRIKTRSNLKKCKSLLSRPSS 343
>BG644364 similar to PIR|E86221|E86 hypothetical protein [imported] -
Arabidopsis thaliana, partial (55%)
Length = 638
Score = 26.6 bits (57), Expect = 5.4
Identities = 16/71 (22%), Positives = 31/71 (43%)
Frame = -3
Query: 79 HLFPFSLRDRASAWFHSLKIGSIISWDQMRKAFLSQFFPHSKTAQLRIKSHSSLKKMVNL 138
H+ P S + W+H++ +SW RK H Q+ + + S+ K +++
Sbjct: 633 HVHPLSSNRLSRLWWHAIIEWQTLSWASNRKRSSRNCCKH-LILQVLLGTMSTKKSNMSV 457
Query: 139 FTMRGSVSKKC 149
M ++S C
Sbjct: 456 LVMAAAISFFC 424
>TC82507
Length = 985
Score = 26.2 bits (56), Expect = 7.1
Identities = 8/20 (40%), Positives = 14/20 (70%)
Frame = -2
Query: 155 IMVLRIGLVSTLFIMVFCFP 174
+ ++R+G S F+ +FCFP
Sbjct: 558 MQIIRVGCSSFFFLFLFCFP 499
>BF519551
Length = 476
Score = 25.8 bits (55), Expect = 9.2
Identities = 11/33 (33%), Positives = 20/33 (60%)
Frame = +2
Query: 12 IISSDEPYDAILYPTVEGNNFEIKSALIYLVQE 44
++SS EP + + V N+F +K +IYL+ +
Sbjct: 305 LVSSIEPGNQVEVVVVVENDFIVKKTIIYLIYD 403
>TC83499
Length = 623
Score = 25.8 bits (55), Expect = 9.2
Identities = 18/64 (28%), Positives = 32/64 (49%)
Frame = +2
Query: 55 PNLHVSIFLRLSGTLKANQEAVRLHLFPFSLRDRASAWFHSLKIGSIISWDQMRKAFLSQ 114
PN+ SI + L +A + +++ FP +L R + L + +I + Q +K L Q
Sbjct: 149 PNICCSILIPLRRQREAGLKRLKVLKFPLALSLRIRGSQNLLLMMVLILF*QDQKHILPQ 328
Query: 115 FFPH 118
FP+
Sbjct: 329 IFPN 340
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.326 0.138 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,606,037
Number of Sequences: 36976
Number of extensions: 75112
Number of successful extensions: 471
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 469
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 471
length of query: 175
length of database: 9,014,727
effective HSP length: 90
effective length of query: 85
effective length of database: 5,686,887
effective search space: 483385395
effective search space used: 483385395
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 55 (25.8 bits)
Medicago: description of AC121241.11


