

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121240.11 + phase: 0 /pseudo
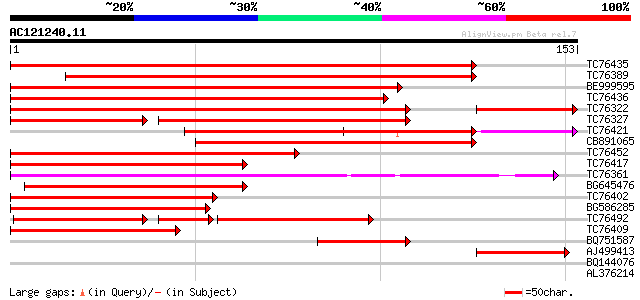
(153 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC76435 homologue to GP|13359451|dbj|BAB33421. putative senescen... 255 4e-69
TC76389 homologue to GP|13359451|dbj|BAB33421. putative senescen... 231 1e-61
BE999595 GP|13359451|db putative senescence-associated protein {... 214 8e-57
TC76436 GP|13359451|dbj|BAB33421. putative senescence-associated... 197 1e-51
TC76322 similar to PIR|T02955|T02955 probable cytochrome P450 mo... 167 2e-42
TC76327 similar to GP|13359451|dbj|BAB33421. putative senescence... 112 4e-41
TC76421 homologue to PIR|T02995|T02995 unspecific monooxygenase ... 157 1e-39
CB891065 similar to PIR|C86216|C86 protein T23G18.6 [imported] -... 157 2e-39
TC76452 homologue to GP|13359451|dbj|BAB33421. putative senescen... 153 2e-38
TC76417 homologue to GP|12049598|emb|CAC19855. mitochondrial suc... 128 1e-30
TC76361 similar to GP|13359451|dbj|BAB33421. putative senescence... 123 3e-29
BG645476 similar to GP|16226228|gb At1g20110/T20H2_10 {Arabidops... 122 6e-29
TC76402 weakly similar to GP|18086561|gb|AAL57705.1 At2g01100/F2... 108 8e-25
BG586285 homologue to GP|13359451|db putative senescence-associa... 102 8e-23
TC76492 similar to GP|13359451|dbj|BAB33421. putative senescence... 59 1e-22
TC76409 similar to GP|13359451|dbj|BAB33421. putative senescence... 84 2e-17
BQ751587 similar to PIR|T02955|T029 probable cytochrome P450 mon... 48 1e-06
AJ499413 similar to PIR|T02995|T029 unspecific monooxygenase (EC... 40 5e-04
BQ144076 similar to GP|7106224|gb| flagelliform silk protein {Ne... 32 0.076
AL376214 homologue to GP|3204132|emb| extensin {Cicer arietinum}... 30 0.29
>TC76435 homologue to GP|13359451|dbj|BAB33421. putative
senescence-associated protein {Pisum sativum}, complete
Length = 1389
Score = 255 bits (652), Expect = 4e-69
Identities = 122/126 (96%), Positives = 124/126 (97%)
Frame = +1
Query: 1 MIGIADIEGSKSNVAMNAWMPQASYPCGNFSDTSSFKFRRSKGSIGHAFTVGIRTGNQNQ 60
MIG ADIEGSKSNVAMNAW+PQASYPCGNFSDTSSFKFRRSKGSIGHAFTV IRTGNQNQ
Sbjct: 637 MIGRADIEGSKSNVAMNAWLPQASYPCGNFSDTSSFKFRRSKGSIGHAFTVRIRTGNQNQ 816
Query: 61 TSFYPFVPHEISVLVKLILGHLRYLLTDVPPQPNSPPDNVFRPDRPTKVGLGSKKRGSAP 120
TSFYPFVPHEISVLV+LILGHLRYLLTDVPPQPNSPPDNVFRPDRPTKVGLGSKKRGSAP
Sbjct: 817 TSFYPFVPHEISVLVELILGHLRYLLTDVPPQPNSPPDNVFRPDRPTKVGLGSKKRGSAP 996
Query: 121 PPIHGI 126
PPIHGI
Sbjct: 997 PPIHGI 1014
>TC76389 homologue to GP|13359451|dbj|BAB33421. putative
senescence-associated protein {Pisum sativum}, partial
(51%)
Length = 808
Score = 231 bits (588), Expect = 1e-61
Identities = 108/111 (97%), Positives = 110/111 (98%)
Frame = +3
Query: 16 MNAWMPQASYPCGNFSDTSSFKFRRSKGSIGHAFTVGIRTGNQNQTSFYPFVPHEISVLV 75
MNAW+PQASYPCGNFSDTSSFKFRRSKGSIGHAFTV IRTGNQNQTSFYPFVPHEISVLV
Sbjct: 375 MNAWLPQASYPCGNFSDTSSFKFRRSKGSIGHAFTVRIRTGNQNQTSFYPFVPHEISVLV 554
Query: 76 KLILGHLRYLLTDVPPQPNSPPDNVFRPDRPTKVGLGSKKRGSAPPPIHGI 126
+LILGHLRYLLTDVPPQPNSPPDNVFRPDRPTKVGLGSKKRGSAPPPIHGI
Sbjct: 555 ELILGHLRYLLTDVPPQPNSPPDNVFRPDRPTKVGLGSKKRGSAPPPIHGI 707
>BE999595 GP|13359451|db putative senescence-associated protein {Pisum
sativum}, partial (37%)
Length = 579
Score = 214 bits (546), Expect = 8e-57
Identities = 102/106 (96%), Positives = 104/106 (97%)
Frame = -2
Query: 1 MIGIADIEGSKSNVAMNAWMPQASYPCGNFSDTSSFKFRRSKGSIGHAFTVGIRTGNQNQ 60
MIG ADIEGSKSNVAMNAW+PQASYPCGNFSDTSSFKFRRSKGSIGHAFTV IRTGNQNQ
Sbjct: 320 MIGRADIEGSKSNVAMNAWLPQASYPCGNFSDTSSFKFRRSKGSIGHAFTVRIRTGNQNQ 141
Query: 61 TSFYPFVPHEISVLVKLILGHLRYLLTDVPPQPNSPPDNVFRPDRP 106
TSFYPFVPHEISVLV+LILGHLRYLLTDVPPQPNSPPDNVFRPDRP
Sbjct: 140 TSFYPFVPHEISVLVELILGHLRYLLTDVPPQPNSPPDNVFRPDRP 3
>TC76436 GP|13359451|dbj|BAB33421. putative senescence-associated protein
{Pisum sativum}, partial (39%)
Length = 774
Score = 197 bits (502), Expect = 1e-51
Identities = 95/102 (93%), Positives = 97/102 (94%)
Frame = +1
Query: 1 MIGIADIEGSKSNVAMNAWMPQASYPCGNFSDTSSFKFRRSKGSIGHAFTVGIRTGNQNQ 60
MIG ADIEGSKSNVAMNAW+PQASYPCGNFSDTSSFKFRRSKGSIGHAFTV IRTGNQNQ
Sbjct: 355 MIGRADIEGSKSNVAMNAWLPQASYPCGNFSDTSSFKFRRSKGSIGHAFTVRIRTGNQNQ 534
Query: 61 TSFYPFVPHEISVLVKLILGHLRYLLTDVPPQPNSPPDNVFR 102
TSFYPFVPHEISVLV+LILGHLRYLLTDVPPQPNSPPD R
Sbjct: 535 TSFYPFVPHEISVLVELILGHLRYLLTDVPPQPNSPPDQTGR 660
>TC76322 similar to PIR|T02955|T02955 probable cytochrome P450 monooxygenase
- maize (fragment), partial (60%)
Length = 3098
Score = 167 bits (422), Expect = 2e-42
Identities = 84/108 (77%), Positives = 87/108 (79%)
Frame = +3
Query: 1 MIGIADIEGSKSNVAMNAWMPQASYPCGNFSDTSSFKFRRSKGSIGHAFTVGIRTGNQNQ 60
MIG ADIEGSKS+VAMNAW PQASYPCGNFS TSS KFR +KGSIGH F V I T NQNQ
Sbjct: 12 MIGRADIEGSKSDVAMNAWPPQASYPCGNFSGTSSLKFRGTKGSIGHTFMVCIHTENQNQ 191
Query: 61 TSFYPFVPHEISVLVKLILGHLRYLLTDVPPQPNSPPDNVFRPDRPTK 108
FYPFV EISVL + LGHLRY LTDVPPQPNSPPDNVF P RP K
Sbjct: 192 GDFYPFVLLEISVLHEPPLGHLRYGLTDVPPQPNSPPDNVFNPGRPAK 335
Score = 50.1 bits (118), Expect = 4e-07
Identities = 21/27 (77%), Positives = 25/27 (91%)
Frame = +3
Query: 127 ITDPFCRLPLPTLFHRPEAVHLGDLMR 153
+TD FCRLPL TLF++PEAVHLGDL+R
Sbjct: 960 VTDLFCRLPLSTLFYQPEAVHLGDLLR 1040
>TC76327 similar to GP|13359451|dbj|BAB33421. putative senescence-associated
protein {Pisum sativum}, partial (64%)
Length = 868
Score = 112 bits (281), Expect(2) = 4e-41
Identities = 54/68 (79%), Positives = 59/68 (86%)
Frame = +2
Query: 41 SKGSIGHAFTVGIRTGNQNQTSFYPFVPHEISVLVKLILGHLRYLLTDVPPQPNSPPDNV 100
+KGSIGHAFTV I T NQNQ SFYPFV HEISVL++LILGHLRYLLTDVPPQPNSPPD V
Sbjct: 416 AKGSIGHAFTVCIHTENQNQMSFYPFVLHEISVLIELILGHLRYLLTDVPPQPNSPPDYV 595
Query: 101 FRPDRPTK 108
F DR ++
Sbjct: 596 FNLDRKSR 619
Score = 71.2 bits (173), Expect(2) = 4e-41
Identities = 33/37 (89%), Positives = 34/37 (91%)
Frame = +3
Query: 1 MIGIADIEGSKSNVAMNAWMPQASYPCGNFSDTSSFK 37
MIG ADIEGSKSNVAM AW+PQASYPCGNFSDTSS K
Sbjct: 294 MIGRADIEGSKSNVAMYAWLPQASYPCGNFSDTSSLK 404
>TC76421 homologue to PIR|T02995|T02995 unspecific monooxygenase (EC
1.14.14.1) - common tobacco, partial (44%)
Length = 5080
Score = 157 bits (398), Expect = 1e-39
Identities = 74/79 (93%), Positives = 76/79 (95%)
Frame = -1
Query: 48 AFTVGIRTGNQNQTSFYPFVPHEISVLVKLILGHLRYLLTDVPPQPNSPPDNVFRPDRPT 107
+ V IRTGNQNQTSFYPFVPHEISVLV+LILGHLRYLLTDVPPQPNSPPDNVFRPDRPT
Sbjct: 4951 SLVVRIRTGNQNQTSFYPFVPHEISVLVELILGHLRYLLTDVPPQPNSPPDNVFRPDRPT 4772
Query: 108 KVGLGSKKRGSAPPPIHGI 126
KVGLGSKKRGSAPPPIHGI
Sbjct: 4771 KVGLGSKKRGSAPPPIHGI 4715
Score = 62.8 bits (151), Expect = 5e-11
Identities = 37/74 (50%), Positives = 39/74 (52%), Gaps = 11/74 (14%)
Frame = -2
Query: 91 PQPNSPPDNVFRP-----------DRPTKVGLGSKKRGSAPPPIHGIITDPFCRLPLPTL 139
P P PP RP +PT LG P + TDPFCRLPLPTL
Sbjct: 4260 PFPVRPPTGTRRPALAAGAAQAVRQQPTGSKLGPPCPALRANPFPEV-TDPFCRLPLPTL 4084
Query: 140 FHRPEAVHLGDLMR 153
FHRPEAVHLGDLMR
Sbjct: 4083 FHRPEAVHLGDLMR 4042
>CB891065 similar to PIR|C86216|C86 protein T23G18.6 [imported] - Arabidopsis
thaliana, partial (33%)
Length = 800
Score = 157 bits (397), Expect = 2e-39
Identities = 74/76 (97%), Positives = 75/76 (98%)
Frame = -2
Query: 51 VGIRTGNQNQTSFYPFVPHEISVLVKLILGHLRYLLTDVPPQPNSPPDNVFRPDRPTKVG 110
V IRTGNQNQTSFYPFVPHEISVLV+LILGHLRYLLTDVPPQPNSPPDNVFRPDRPTKVG
Sbjct: 370 VRIRTGNQNQTSFYPFVPHEISVLVELILGHLRYLLTDVPPQPNSPPDNVFRPDRPTKVG 191
Query: 111 LGSKKRGSAPPPIHGI 126
LGSKKRGSAPPPIHGI
Sbjct: 190 LGSKKRGSAPPPIHGI 143
>TC76452 homologue to GP|13359451|dbj|BAB33421. putative senescence-associated
protein {Pisum sativum}, partial (29%)
Length = 1664
Score = 153 bits (387), Expect = 2e-38
Identities = 74/78 (94%), Positives = 76/78 (96%)
Frame = -3
Query: 1 MIGIADIEGSKSNVAMNAWMPQASYPCGNFSDTSSFKFRRSKGSIGHAFTVGIRTGNQNQ 60
MIG ADIEGSKSNVAMNAW+PQASYPCGNFSDTSSFKFRRSKGSIGHAFTV IRTGNQNQ
Sbjct: 1044 MIGRADIEGSKSNVAMNAWLPQASYPCGNFSDTSSFKFRRSKGSIGHAFTVRIRTGNQNQ 865
Query: 61 TSFYPFVPHEISVLVKLI 78
TSFYPFVPHEISVLV+LI
Sbjct: 864 TSFYPFVPHEISVLVELI 811
>TC76417 homologue to GP|12049598|emb|CAC19855. mitochondrial succinate
dehydrogenase iron-sulphur subunit {Arabidopsis
thaliana}, partial (81%)
Length = 1200
Score = 128 bits (321), Expect = 1e-30
Identities = 61/64 (95%), Positives = 62/64 (96%)
Frame = -1
Query: 1 MIGIADIEGSKSNVAMNAWMPQASYPCGNFSDTSSFKFRRSKGSIGHAFTVGIRTGNQNQ 60
MIG ADIEGSKSNVAMNAW+PQASYPCGNFSDTSSFKFRRSKGSIGHAFTV IRTGNQNQ
Sbjct: 192 MIGRADIEGSKSNVAMNAWLPQASYPCGNFSDTSSFKFRRSKGSIGHAFTVRIRTGNQNQ 13
Query: 61 TSFY 64
TSFY
Sbjct: 12 TSFY 1
>TC76361 similar to GP|13359451|dbj|BAB33421. putative senescence-associated
protein {Pisum sativum}, partial (47%)
Length = 570
Score = 123 bits (308), Expect = 3e-29
Identities = 77/148 (52%), Positives = 87/148 (58%)
Frame = +1
Query: 1 MIGIADIEGSKSNVAMNAWMPQASYPCGNFSDTSSFKFRRSKGSIGHAFTVGIRTGNQNQ 60
MIG ADIEGSKS+VAM AW PQASYPCGNFS TSS KFR +KGSIGH F V I T NQNQ
Sbjct: 10 MIGRADIEGSKSDVAMYAWPPQASYPCGNFSGTSSLKFRGTKGSIGHTFMVCIHTENQNQ 189
Query: 61 TSFYPFVPHEISVLVKLILGHLRYLLTDVPPQPNSPPDNVFRPDRPTKVGLGSKKRGSAP 120
FYPFV EISVL + LGHLRY P P +P + G ++K P
Sbjct: 190 GDFYPFVLLEISVLHEPPLGHLRYGFRCAAP-AKLPT*QCLQPG-SAREGP*NQKLDVNP 363
Query: 121 PPIHGIITDPFCRLPLPTLFHRPEAVHL 148
P+H I +FHR + HL
Sbjct: 364 APLH*ISKKTIG----VVVFHRRRSSHL 435
>BG645476 similar to GP|16226228|gb At1g20110/T20H2_10 {Arabidopsis
thaliana}, partial (28%)
Length = 690
Score = 122 bits (306), Expect = 6e-29
Identities = 57/60 (95%), Positives = 59/60 (98%)
Frame = -2
Query: 5 ADIEGSKSNVAMNAWMPQASYPCGNFSDTSSFKFRRSKGSIGHAFTVGIRTGNQNQTSFY 64
+DIEGSKSNVAMNAW+PQASYPCGNFSDTSSFKFRRSKGSIGHAFTV IRTGNQNQTSFY
Sbjct: 182 SDIEGSKSNVAMNAWLPQASYPCGNFSDTSSFKFRRSKGSIGHAFTVRIRTGNQNQTSFY 3
>TC76402 weakly similar to GP|18086561|gb|AAL57705.1 At2g01100/F23H14.7
{Arabidopsis thaliana}, partial (43%)
Length = 1449
Score = 108 bits (270), Expect = 8e-25
Identities = 52/56 (92%), Positives = 53/56 (93%)
Frame = -1
Query: 1 MIGIADIEGSKSNVAMNAWMPQASYPCGNFSDTSSFKFRRSKGSIGHAFTVGIRTG 56
MIG ADIEGSKSNVAMNAW+PQASYPCGNFSDTSSFKFRRSKGS GHAFTV IRTG
Sbjct: 168 MIGRADIEGSKSNVAMNAWLPQASYPCGNFSDTSSFKFRRSKGSRGHAFTVRIRTG 1
>BG586285 homologue to GP|13359451|db putative senescence-associated protein
{Pisum sativum}, partial (45%)
Length = 704
Score = 102 bits (253), Expect = 8e-23
Identities = 49/54 (90%), Positives = 50/54 (91%)
Frame = +3
Query: 1 MIGIADIEGSKSNVAMNAWMPQASYPCGNFSDTSSFKFRRSKGSIGHAFTVGIR 54
MIG ADIE SKSNVAMNAW+PQA YPCGNFSDTSSFKFRRSKGSIGHAFTV IR
Sbjct: 543 MIGRADIERSKSNVAMNAWLPQARYPCGNFSDTSSFKFRRSKGSIGHAFTVRIR 704
>TC76492 similar to GP|13359451|dbj|BAB33421. putative senescence-associated
protein {Pisum sativum}, partial (35%)
Length = 829
Score = 59.3 bits (142), Expect(3) = 1e-22
Identities = 28/42 (66%), Positives = 33/42 (77%)
Frame = +3
Query: 57 NQNQTSFYPFVPHEISVLVKLILGHLRYLLTDVPPQPNSPPD 98
N+ + SFYPFV H+ISVLV+LI GHL Y LT V PQPNSP +
Sbjct: 582 NKIKCSFYPFVQHKISVLVELIFGHLCYHLTHVTPQPNSPSE 707
Score = 55.8 bits (133), Expect(3) = 1e-22
Identities = 26/36 (72%), Positives = 30/36 (83%)
Frame = +2
Query: 2 IGIADIEGSKSNVAMNAWMPQASYPCGNFSDTSSFK 37
+G ADIE SKS+VA+NAW PQASYP GN+SDTS K
Sbjct: 383 VGRADIEESKSDVALNAWPPQASYPYGNYSDTSIAK 490
Score = 26.6 bits (57), Expect(3) = 1e-22
Identities = 12/15 (80%), Positives = 13/15 (86%)
Frame = +1
Query: 41 SKGSIGHAFTVGIRT 55
+KGSIGHAFTV I T
Sbjct: 529 NKGSIGHAFTVCIPT 573
>TC76409 similar to GP|13359451|dbj|BAB33421. putative senescence-associated
protein {Pisum sativum}, partial (40%)
Length = 614
Score = 84.3 bits (207), Expect = 2e-17
Identities = 40/46 (86%), Positives = 41/46 (88%)
Frame = -3
Query: 1 MIGIADIEGSKSNVAMNAWMPQASYPCGNFSDTSSFKFRRSKGSIG 46
MIG ADIEGSKSNVAMNAW+PQASYPCGNFSDTSS R SKGSIG
Sbjct: 138 MIGRADIEGSKSNVAMNAWLPQASYPCGNFSDTSSLTLRGSKGSIG 1
>BQ751587 similar to PIR|T02955|T029 probable cytochrome P450 monooxygenase -
maize (fragment), partial (20%)
Length = 398
Score = 48.1 bits (113), Expect = 1e-06
Identities = 20/25 (80%), Positives = 20/25 (80%)
Frame = -1
Query: 84 YLLTDVPPQPNSPPDNVFRPDRPTK 108
Y TDVPPQPNSPPDNVF P RP K
Sbjct: 275 YGFTDVPPQPNSPPDNVFNPGRPAK 201
>AJ499413 similar to PIR|T02995|T029 unspecific monooxygenase (EC 1.14.14.1)
- common tobacco, partial (5%)
Length = 365
Score = 39.7 bits (91), Expect = 5e-04
Identities = 18/25 (72%), Positives = 21/25 (84%)
Frame = -2
Query: 127 ITDPFCRLPLPTLFHRPEAVHLGDL 151
+TD FCRLPL TLF++ EAVHLG L
Sbjct: 88 VTDLFCRLPLLTLFYQLEAVHLGRL 14
>BQ144076 similar to GP|7106224|gb| flagelliform silk protein {Nephila
clavipes}, partial (2%)
Length = 1163
Score = 32.3 bits (72), Expect = 0.076
Identities = 20/48 (41%), Positives = 22/48 (45%)
Frame = -2
Query: 90 PPQPNSPPDNVFRPDRPTKVGLGSKKRGSAPPPIHGIITDPFCRLPLP 137
PP P +PP RP P VG G R + PPP G P R P P
Sbjct: 154 PPPPAAPPTP--RPPGPAPVGPGGGGRPTPPPPNRGTAGTP--RPPTP 23
>AL376214 homologue to GP|3204132|emb| extensin {Cicer arietinum}, partial
(48%)
Length = 362
Score = 30.4 bits (67), Expect = 0.29
Identities = 15/56 (26%), Positives = 26/56 (45%)
Frame = +3
Query: 68 PHEISVLVKLILGHLRYLLTDVPPQPNSPPDNVFRPDRPTKVGLGSKKRGSAPPPI 123
PH + + +K++L L + PP + PP + P P+ + S PPP+
Sbjct: 57 PHHLLITIKVLLHLLTIM*VPPPPTSSPPPYHYTSPPPPSPAPAPTYIYKSPPPPM 224
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.141 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,260,670
Number of Sequences: 36976
Number of extensions: 89370
Number of successful extensions: 717
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 640
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 702
length of query: 153
length of database: 9,014,727
effective HSP length: 88
effective length of query: 65
effective length of database: 5,760,839
effective search space: 374454535
effective search space used: 374454535
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 54 (25.4 bits)
Medicago: description of AC121240.11


