

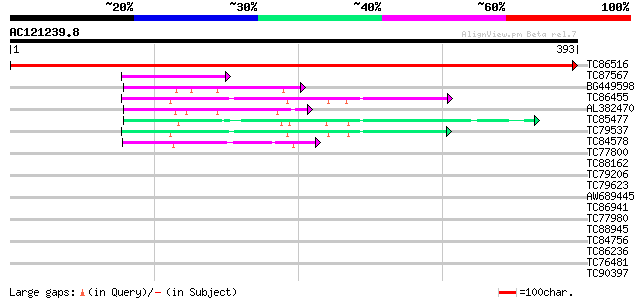
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121239.8 + phase: 0
(393 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86516 homologue to GP|18087583|gb|AAL58922.1 AT4g35250/F23E12_... 794 0.0
TC87567 similar to GP|17065112|gb|AAL32710.1 Unknown protein {Ar... 56 3e-08
BG449598 weakly similar to GP|4336887|gb| sophorol reductase {Pi... 54 1e-07
TC86455 similar to SP|P52581|IFRI_LUPAL Isoflavone reductase hom... 47 1e-05
AL382470 similar to PIR|S66262|S66 vestitone reductase - alfalfa... 47 1e-05
TC85477 isoflavone reductase 46 3e-05
TC79537 similar to SP|P52581|IFRI_LUPAL Isoflavone reductase hom... 45 6e-05
TC84578 43 2e-04
TC77800 similar to PIR|T02532|T02532 hypothetical protein At2g37... 40 0.001
TC88162 similar to GP|21616072|emb|CAC87810. Tic62 protein {Pisu... 40 0.002
TC79206 similar to GP|20339364|gb|AAM19355.1 UOS1 {Pisum sativum... 40 0.002
TC79623 similar to GP|20339364|gb|AAM19355.1 UOS1 {Pisum sativum... 37 0.010
AW689445 similar to SP|P52581|IFRI_ Isoflavone reductase homolog... 36 0.029
TC86941 similar to GP|15294290|gb|AAK95322.1 AT5g02240/T7H20_290... 35 0.038
TC77980 similar to PIR|T12970|T12970 hypothetical protein T6H20.... 32 0.32
TC88945 similar to PIR|T05987|T05987 hypothetical protein F17M5.... 30 1.6
TC84756 30 1.6
TC86236 similar to PIR|T05311|T05311 sulfolipid biosynthesis pro... 28 4.7
TC76481 similar to PIR|G84677|G84677 probable dTDP-glucose 4-6-d... 28 8.0
TC90397 similar to GP|15010740|gb|AAK74029.1 AT5g59730/mth12_130... 28 8.0
>TC86516 homologue to GP|18087583|gb|AAL58922.1 AT4g35250/F23E12_190
{Arabidopsis thaliana}, partial (83%)
Length = 1499
Score = 794 bits (2050), Expect = 0.0
Identities = 393/393 (100%), Positives = 393/393 (100%)
Frame = +2
Query: 1 MKMATLMRLPTATPGRVVTRTREAFHALSPSPSPSPPHFVEFCKGGRRGRRIIRVGVKCN 60
MKMATLMRLPTATPGRVVTRTREAFHALSPSPSPSPPHFVEFCKGGRRGRRIIRVGVKCN
Sbjct: 95 MKMATLMRLPTATPGRVVTRTREAFHALSPSPSPSPPHFVEFCKGGRRGRRIIRVGVKCN 274
Query: 61 SNSERAVNLGPGTPVRPTSILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRD 120
SNSERAVNLGPGTPVRPTSILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRD
Sbjct: 275 SNSERAVNLGPGTPVRPTSILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRD 454
Query: 121 WGATVVNADLSKPETIPATLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQK 180
WGATVVNADLSKPETIPATLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQK
Sbjct: 455 WGATVVNADLSKPETIPATLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQK 634
Query: 181 YVFYSIHNCDKHPEVPLMEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKS 240
YVFYSIHNCDKHPEVPLMEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKS
Sbjct: 635 YVFYSIHNCDKHPEVPLMEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKS 814
Query: 241 VWGTDAPTRIAYMDTQDIARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQD 300
VWGTDAPTRIAYMDTQDIARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQD
Sbjct: 815 VWGTDAPTRIAYMDTQDIARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQD 994
Query: 301 ANVTTVPVSVLRLTRQLTRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKD 360
ANVTTVPVSVLRLTRQLTRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKD
Sbjct: 995 ANVTTVPVSVLRLTRQLTRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKD 1174
Query: 361 IITLEKYLQDYFTNILKKLKDLKAQSKQSDIFF 393
IITLEKYLQDYFTNILKKLKDLKAQSKQSDIFF
Sbjct: 1175IITLEKYLQDYFTNILKKLKDLKAQSKQSDIFF 1273
Score = 29.3 bits (64), Expect = 2.7
Identities = 17/38 (44%), Positives = 21/38 (54%)
Frame = -2
Query: 24 AFHALSPSPSPSPPHFVEFCKGGRRGRRIIRVGVKCNS 61
AFH+ S +PS S F + K RR RR R +KC S
Sbjct: 271 AFHSHSNNPSSSSSTFAKLNKMRRRRRRRRRKSMKCLS 158
>TC87567 similar to GP|17065112|gb|AAL32710.1 Unknown protein {Arabidopsis
thaliana}, partial (82%)
Length = 2117
Score = 55.8 bits (133), Expect = 3e-08
Identities = 29/76 (38%), Positives = 42/76 (55%)
Frame = +2
Query: 78 TSILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPETIP 137
T++LVVGAT +GR +VR+ + GY V+ LVR L +V D+ P T+
Sbjct: 557 TTVLVVGATSRIGRIVVRKLMLRGYSVKALVRKADEEVVELLPRSVEIVIGDVGDPNTVK 736
Query: 138 ATLVGVHTVIDCATGR 153
A + G + +I CAT R
Sbjct: 737 AAVEGCNKIIYCATAR 784
>BG449598 weakly similar to GP|4336887|gb| sophorol reductase {Pisum
sativum}, partial (53%)
Length = 650
Score = 53.5 bits (127), Expect = 1e-07
Identities = 41/143 (28%), Positives = 64/143 (44%), Gaps = 17/143 (11%)
Frame = +2
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAP---ADFLRDWGAT-----VVNADLS 131
+ V G TG +G I++R L++GY V VR P FL D + NADLS
Sbjct: 125 VCVTGGTGFIGSWIIKRLLEDGYTVNTTVRSNPGQKKDVSFLTDLPNASQKLQIFNADLS 304
Query: 132 KPETIPATLVG------VHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKYVFYS 185
PE+ A + G T +D PEE + +G + +++ K K V Y+
Sbjct: 305 NPESFNAAIEGCIGVFHTATPVDFELKEPEEIVIKRTIDGALGILKACKNSKTVKRVVYT 484
Query: 186 IHN---CDKHPEVPLMEIKYCTE 205
C ++ EV +M+ Y ++
Sbjct: 485 SSASAVCMQNKEVEVMDESYWSD 553
>TC86455 similar to SP|P52581|IFRI_LUPAL Isoflavone reductase homolog (EC
1.3.1.-). [White lupine] {Lupinus albus}, complete
Length = 1194
Score = 47.4 bits (111), Expect = 1e-05
Identities = 55/250 (22%), Positives = 103/250 (41%), Gaps = 20/250 (8%)
Frame = +3
Query: 78 TSILVVGATGTLGRQIVRRALDEGYDVRCLVRP-------RPAPADFLRDWGATVVNADL 130
+ +LVVG TG +GR+IV+ +L++G++ + RP + + GA +V A
Sbjct: 51 SKVLVVGGTGYIGRRIVKASLEQGHETYVIQRPELGLQIEKLQRLLSFKKQGAHIVEASF 230
Query: 131 SKPETIPATLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCD 190
S +++ + V VI +G I++ ++ L+ K G K S D
Sbjct: 231 SDHKSLVDAIKKVDVVISAISG---VHIRSHSIGLQLKLVDAIKEAGNIKRFLPSEFGLD 401
Query: 191 K-------HPEVPLMEIKYCTENFLRDSGLNHIVIR---LCGFMQGLIGQYA--VPILEE 238
P + K + ++ + I G+ G + Q VP ++
Sbjct: 402 PARMGHALEPGRVTFDDKMAVRKAIEEANIPFTYISANLFAGYFAGSLSQMGSFVPPRDK 581
Query: 239 KSVWGTDAPTRIAYMDTQDIARLTFIALRNEKINGKLLTFAGPRAWTTQ-EVITLCERLA 297
++G D + ++D D+A T + + + K L +Q E+I + E+L
Sbjct: 582 VHLFG-DGKHKAIFLDEYDVATYTIKTIDDPRTLNKTLYLRPQENILSQGELIGIWEKLI 758
Query: 298 GQDANVTTVP 307
G+D T +P
Sbjct: 759 GKDLEKTYIP 788
>AL382470 similar to PIR|S66262|S66 vestitone reductase - alfalfa, partial
(48%)
Length = 550
Score = 47.4 bits (111), Expect = 1e-05
Identities = 42/152 (27%), Positives = 67/152 (43%), Gaps = 21/152 (13%)
Frame = -3
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPA----PADFLRDW-----GATVVNADL 130
+ V G TG +G I++R L++GY V VR PA +L + + NADL
Sbjct: 470 VCVTGGTGFIGSWIIKRLLEDGYTVNTTVRSNPAGQKKDVSYLTNLPNASQNLQIFNADL 291
Query: 131 SKPETIPATLVG------VHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKYVFY 184
PE+ A + G T ID EE + +G + +++ K K V Y
Sbjct: 290 CNPESFDAAIEGCIGIFHTATPIDFEENEREEIVTKRTIDGALGILKACKNSKTVKRVIY 111
Query: 185 -----SIHNCDKHPEVPLMEIKYCTE-NFLRD 210
+++ DK +V M+ Y ++ N LR+
Sbjct: 110 TSSASAVYMQDKEEDV--MDESYWSDVNILRN 21
>TC85477 isoflavone reductase
Length = 1354
Score = 45.8 bits (107), Expect = 3e-05
Identities = 75/318 (23%), Positives = 121/318 (37%), Gaps = 30/318 (9%)
Frame = +3
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPA------------------DFLRDW 121
IL++G TG +GR IV ++ G LVR P D +
Sbjct: 222 ILILGPTGAIGRHIVWASIKAGNPTYALVRKTPGNVNKPKLITAANPETKEELIDNYQSL 401
Query: 122 GATVVNADLSKPETIPATLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKY 181
G ++ D++ ET+ + V VI CA GR + E +V +I+ K G K
Sbjct: 402 GVILLEGDINDHETLVKAIKQVDIVI-CAAGR-------LLIEDQVKIIKAIKEAGNVKK 557
Query: 182 VFYSIH--NCDKH----PEVPLMEIKYCTENFLRDSGLNHIVI---RLCGFMQGLIGQYA 232
F S + D+H P + E K + G+ + + G+ + Q
Sbjct: 558 FFPSEFGLDVDRHEAVEPVRQVFEEKASIRRVIEAEGVPYTYLCCHAFTGYFLRNLAQLD 737
Query: 233 V--PILEEKSVWGTDAPTRIAYMDTQDIARLTFIALRNEKINGKLLTFAGPRAWTTQ-EV 289
V P ++ + G D + AY+ D+ T A + K + P+ + TQ EV
Sbjct: 738 VTDPPRDKVVILG-DGNVKGAYVTEADVGTFTIKAANDPNTLNKAVHIRLPKNYLTQNEV 914
Query: 290 ITLCERLAGQDANVTTVPVSVLRLTRQLTRFFEWTNDVADRLAFSEVLTSDTVFSVPMAE 349
I+L E+ G+ T V + Q + F ++ L S+ + D V+ +
Sbjct: 915 ISLWEKKIGKTLEKTYVSEEQVLKDIQESSF---PHNYLLALYHSQQIKGDAVYEID--- 1076
Query: 350 TYNLLGVDTKDIITLEKY 367
TKDI E Y
Sbjct: 1077-------PTKDIEASEAY 1109
>TC79537 similar to SP|P52581|IFRI_LUPAL Isoflavone reductase homolog (EC
1.3.1.-). [White lupine] {Lupinus albus}, complete
Length = 1069
Score = 44.7 bits (104), Expect = 6e-05
Identities = 54/249 (21%), Positives = 101/249 (39%), Gaps = 20/249 (8%)
Frame = +1
Query: 78 TSILVVGATGTLGRQIVRRALDEGYDVRCLVRP-------RPAPADFLRDWGATVVNADL 130
+ +LVVG TG +GR+IV+ +L++G++ L RP + + GA +V
Sbjct: 43 SKVLVVGGTGYIGRRIVKASLEQGHETYVLQRPDIGLETEKVQMLLSFKKLGAHLVEGSF 222
Query: 131 SKPETIPATLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCD 190
S +++ + V VI +G ++ + ++ LI+ K G K S D
Sbjct: 223 SNHQSLVDAVKLVDVVICTMSG---VHFRSHNLMLQLKLIEAIKDAGNVKRFLPSEFGMD 393
Query: 191 K-------HPEVPLMEIKYCTENFLRDSGLNHIVIR---LCGFMQGLIGQYAV--PILEE 238
P + K + D+ + I + G + Q P ++
Sbjct: 394 PALMGHALEPGRVTFDEKMTIRKTIEDANIPFTYISANCFAAYFAGNLSQMGTLFPPRDK 573
Query: 239 KSVWGTDAPTRIAYMDTQDIARLTFIALRNEKINGKLLTFAGPRAWTTQ-EVITLCERLA 297
++G D ++ YMD D+A T + + + K + P TQ E+I E++
Sbjct: 574 VVLYG-DGNVKVVYMDEDDVATYTIKTIDDPRTLNKTIYIRPPENILTQRELIEKWEKII 750
Query: 298 GQDANVTTV 306
G+ +T+
Sbjct: 751 GKQLEKSTI 777
>TC84578
Length = 792
Score = 42.7 bits (99), Expect = 2e-04
Identities = 34/143 (23%), Positives = 69/143 (47%), Gaps = 6/143 (4%)
Frame = +3
Query: 79 SILVVGATGTLGRQIVRRALDEGYDVRCLVRPRP----APADFLRDWGATVVNADLSKPE 134
++++ GATGTLG I L++G + R +P ++ L GA +V D ++
Sbjct: 114 TVVIAGATGTLGYPITEAFLNDGSYKIKIFRKKPESKNEKSELLASKGAEIVYVDYTQKG 293
Query: 135 TIPATLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKYVFYSIHNCDKHPE 194
+ L G VI + + + + +++L++ AK +G+++++ S + P+
Sbjct: 294 DLVKALKGTDVVISALS---PKKMGGDFYSTQLSLLEAAKEVGVKRFI-PSEFGGEYGPD 461
Query: 195 V--PLMEIKYCTENFLRDSGLNH 215
+ P +E+K L SGL +
Sbjct: 462 IAHPFIEVKIKFREVLEKSGLEY 530
>TC77800 similar to PIR|T02532|T02532 hypothetical protein At2g37660
[imported] - Arabidopsis thaliana, partial (73%)
Length = 1379
Score = 40.4 bits (93), Expect = 0.001
Identities = 62/234 (26%), Positives = 95/234 (40%), Gaps = 32/234 (13%)
Frame = +1
Query: 78 TSILVVGATGTLGRQIVRRALDEG---YDVRCLVRPRPAPADFLRDWGAT--VVNADLSK 132
+++LV GA G G QIV + L E Y R LVR + GA V D+
Sbjct: 415 STVLVTGAGGRTG-QIVYKKLKERPNEYIARGLVRSEESKQKI----GAADDVFIGDIRD 579
Query: 133 PETIPATLVGVHTVIDCATG--------------RPEEPIKT------VDWEGKVALIQC 172
E++ + G+ +I +G RPE + VDW G+ I
Sbjct: 580 TESLAPAIQGIDALIILTSGVPLMKPGFDPTQGKRPEFYFEDGAYPEQVDWIGQKNQIDA 759
Query: 173 AKAMGIQKYVFY-SIHNCD-KHP-----EVPLMEIKYCTENFLRDSGLNHIVIRLCGFMQ 225
AKA G+++ V S+ D HP + ++ K E +L DSG+ + +IR G
Sbjct: 760 AKAAGVKQIVLVGSMGGTDLNHPLNSLGDGNILVWKRKAEQYLADSGIPYTIIRAGGLQD 939
Query: 226 GLIGQYAVPILEEKSVWGTDAPTRIAYMDTQDIARLTFIALRNEKINGKLLTFA 279
G + I ++ + TD T + D+A + AL E+ K A
Sbjct: 940 KEGGIRELVIGKDDELLKTDIRT----IARPDVAEVCLQALNFEEAQFKAFDLA 1089
>TC88162 similar to GP|21616072|emb|CAC87810. Tic62 protein {Pisum sativum},
partial (36%)
Length = 634
Score = 39.7 bits (91), Expect = 0.002
Identities = 42/148 (28%), Positives = 57/148 (38%), Gaps = 27/148 (18%)
Frame = +2
Query: 38 HFVEF------CKGGRRGRRIIRVGVKCNSNSERAVNLGPGTPVRPTS-----ILVVGAT 86
HF+ + CK R IIR V ++ S A G P + S + V GAT
Sbjct: 128 HFINYPFTTTPCKNRIRSN-IIRAQVSGSTKSSTA----EGIPEKKDSKDDNLVFVAGAT 292
Query: 87 GTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGAT----------------VVNADL 130
G +G + VR + G+ VR VR ++ +V DL
Sbjct: 293 GKVGSRTVRELIKLGFKVRAGVRSAQRAGSLVQSVKQLKLDGTSGGSEAVEKLEIVECDL 472
Query: 131 SKPETIPATLVGVHTVIDCATGRPEEPI 158
KPE I + L TVI C G E+ I
Sbjct: 473 EKPEQIKSALGNASTVI-CTIGASEKEI 553
>TC79206 similar to GP|20339364|gb|AAM19355.1 UOS1 {Pisum sativum}, partial
(40%)
Length = 842
Score = 39.7 bits (91), Expect = 0.002
Identities = 37/119 (31%), Positives = 51/119 (42%), Gaps = 1/119 (0%)
Frame = +1
Query: 34 PSPPHFVEFCKGGRRGRRIIRVGVKCNSNSERAVNLGPGTPVRPTSILVVGATGTLGRQI 93
PSP F +F G + +S SE ++G V LV GATG +GR++
Sbjct: 295 PSPAKFFDFLVGK----------LSSSSTSESVNSMGTSDIV-----LVAGATGGVGRRV 429
Query: 94 VRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPET-IPATLVGVHTVIDCAT 151
V +G VR LVR L +V D++K T IP GV VI+ +
Sbjct: 430 VDELRKKGIPVRVLVRNEEKARKMLGS-DVDLVIGDITKDSTLIPEYFKGVKKVINAVS 603
>TC79623 similar to GP|20339364|gb|AAM19355.1 UOS1 {Pisum sativum}, partial
(32%)
Length = 691
Score = 37.4 bits (85), Expect = 0.010
Identities = 31/98 (31%), Positives = 47/98 (47%), Gaps = 1/98 (1%)
Frame = +1
Query: 55 VGVKCNSNSERAVNLGPGTPVRPTSILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAP 114
VG +S++ +VN + + +LV GATG +GR++V +G VR LVR
Sbjct: 322 VGKLSSSSTSESVNSMGTSDI----VLVAGATGGVGRRVVDELRKKGIPVRVLVRNEEKA 489
Query: 115 ADFLRDWGATVVNADLSKPET-IPATLVGVHTVIDCAT 151
L +V D++K T IP GV VI+ +
Sbjct: 490 RKMLGS-DVDLVIGDITKDSTLIPEYFKGVKKVINAVS 600
>AW689445 similar to SP|P52581|IFRI_ Isoflavone reductase homolog (EC
1.3.1.-). [White lupine] {Lupinus albus}, partial (12%)
Length = 207
Score = 35.8 bits (81), Expect = 0.029
Identities = 15/33 (45%), Positives = 24/33 (72%)
Frame = +1
Query: 78 TSILVVGATGTLGRQIVRRALDEGYDVRCLVRP 110
+ +LVVG TG +GR+IV+ +L +G++ L RP
Sbjct: 22 SKVLVVGGTGYIGRRIVKASLXQGHETYVLQRP 120
>TC86941 similar to GP|15294290|gb|AAK95322.1 AT5g02240/T7H20_290
{Arabidopsis thaliana}, partial (98%)
Length = 1121
Score = 35.4 bits (80), Expect = 0.038
Identities = 57/234 (24%), Positives = 90/234 (38%), Gaps = 32/234 (13%)
Frame = +2
Query: 78 TSILVVGATGTLGRQIVRRALDEGYD---VRCLVRPRPAPADFLRDWGAT-VVNADLSKP 133
T++LV GA G G QIV + L E D R LVR + GA + D+
Sbjct: 119 TTVLVTGAGGRTG-QIVYKKLKEKRDQYIARGLVRSEESKQKI---GGADDIFLGDIRNA 286
Query: 134 ETIPATLVGVHTVIDCATGRPE-----EPIK---------------TVDWEGKVALIQCA 173
E+I + G +I + P+ +P K VDW G+ I A
Sbjct: 287 ESIVPAIQGTDALIILTSAVPQMKPGFDPTKGGRPEFYFDDGAYPEQVDWIGQKNQIDAA 466
Query: 174 KAMGIQKYVFYSIHNCDKHPEVPLMEI--------KYCTENFLRDSGLNHIVIRLCGFMQ 225
KA G++ V +P PL + K E +L +SG+ + +IR G +
Sbjct: 467 KAAGVKHIVLVGSMG-GTNPNHPLNSLGNGNILVWKRKAEEYLSNSGVPYTIIRPGGLLD 643
Query: 226 GLIGQYAVPILEEKSVWGTDAPTRIAYMDTQDIARLTFIALRNEKINGKLLTFA 279
G + + ++ + T+ T + D+A + L E+ K A
Sbjct: 644 KEGGVRELIVGKDDELLQTETKT----IPRADVAEVCVQVLNYEETKLKAFDLA 793
>TC77980 similar to PIR|T12970|T12970 hypothetical protein T6H20.190 -
Arabidopsis thaliana, partial (42%)
Length = 1061
Score = 32.3 bits (72), Expect = 0.32
Identities = 16/37 (43%), Positives = 23/37 (61%), Gaps = 3/37 (8%)
Frame = +2
Query: 72 GTPVR---PTSILVVGATGTLGRQIVRRALDEGYDVR 105
G+P R P+++ V GATG G +I + L EG+ VR
Sbjct: 377 GSPTRKKDPSTVFVAGATGQAGIRIAQTLLREGFSVR 487
>TC88945 similar to PIR|T05987|T05987 hypothetical protein F17M5.120 -
Arabidopsis thaliana, partial (94%)
Length = 1345
Score = 30.0 bits (66), Expect = 1.6
Identities = 13/35 (37%), Positives = 21/35 (59%)
Frame = +3
Query: 76 RPTSILVVGATGTLGRQIVRRALDEGYDVRCLVRP 110
R ++V GA+G LG ++ +GY V+ +VRP
Sbjct: 99 RMKKVVVTGASGYLGGKLCNSLHRQGYSVKVIVRP 203
>TC84756
Length = 368
Score = 30.0 bits (66), Expect = 1.6
Identities = 14/38 (36%), Positives = 23/38 (59%)
Frame = +3
Query: 355 GVDTKDIITLEKYLQDYFTNILKKLKDLKAQSKQSDIF 392
G+DT+ TL Y + YF+N KK+ + K ++ S+ F
Sbjct: 210 GLDTESTDTLVSYFKSYFSNKGKKVPENKKSNRYSNNF 323
>TC86236 similar to PIR|T05311|T05311 sulfolipid biosynthesis protein SQD1
chloroplast [validated] - Arabidopsis thaliana, partial
(83%)
Length = 1918
Score = 28.5 bits (62), Expect = 4.7
Identities = 12/45 (26%), Positives = 21/45 (46%)
Frame = +1
Query: 60 NSNSERAVNLGPGTPVRPTSILVVGATGTLGRQIVRRALDEGYDV 104
++ E V G P +P ++V+G G G ++GY+V
Sbjct: 358 STGQEAPVQTSSGDPFKPKRVMVIGGDGYCGWATALHLSNKGYEV 492
>TC76481 similar to PIR|G84677|G84677 probable dTDP-glucose 4-6-dehydratase
[imported] - Arabidopsis thaliana, partial (98%)
Length = 1811
Score = 27.7 bits (60), Expect = 8.0
Identities = 9/29 (31%), Positives = 18/29 (62%)
Frame = +3
Query: 72 GTPVRPTSILVVGATGTLGRQIVRRALDE 100
G P++P +I ++GA G +G + + + E
Sbjct: 183 GNPIKPLTICMIGAGGFIGSHLCEKLMSE 269
>TC90397 similar to GP|15010740|gb|AAK74029.1 AT5g59730/mth12_130 {Arabidopsis
thaliana}, partial (22%)
Length = 1162
Score = 27.7 bits (60), Expect = 8.0
Identities = 16/45 (35%), Positives = 25/45 (55%)
Frame = -1
Query: 318 TRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDII 362
T FFE + +AD +F+ VLT+ FS+ + L VD+ + I
Sbjct: 1018 TCFFEESF*IADSNSFNIVLTASASFSIELMACKRTLEVDSNEKI 884
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.138 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,237,291
Number of Sequences: 36976
Number of extensions: 159635
Number of successful extensions: 1225
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 1060
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1222
length of query: 393
length of database: 9,014,727
effective HSP length: 98
effective length of query: 295
effective length of database: 5,391,079
effective search space: 1590368305
effective search space used: 1590368305
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC121239.8


