

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121239.3 + phase: 0 /pseudo
(616 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
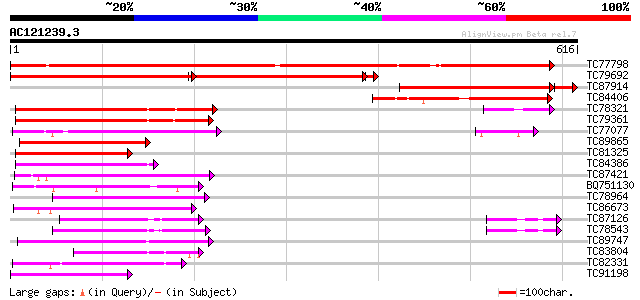
Score E
Sequences producing significant alignments: (bits) Value
TC77798 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_14... 844 0.0
TC79692 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_14... 392 0.0
TC87914 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_14... 338 e-104
TC84406 similar to GP|12039327|gb|AAG46115.1 putative sugar tran... 185 5e-47
TC78321 similar to GP|21689841|gb|AAM67564.1 unknown protein {Ar... 154 7e-38
TC79361 similar to GP|17380890|gb|AAL36257.1 putative membrane t... 151 8e-37
TC77077 similar to GP|9293909|dbj|BAB01812.1 sugar transporter p... 136 3e-32
TC89865 similar to GP|17380890|gb|AAL36257.1 putative membrane t... 109 3e-24
TC81325 similar to GP|17380890|gb|AAL36257.1 putative membrane t... 105 7e-23
TC84386 similar to GP|21689841|gb|AAM67564.1 unknown protein {Ar... 104 1e-22
TC87421 sugar transporter 93 3e-19
BQ751130 weakly similar to GP|15289796|dbj putative monosacchari... 92 6e-19
TC78964 weakly similar to PIR|T00450|T00450 probable monosacchar... 92 6e-19
TC86673 weakly similar to PIR|S25015|S25015 monosaccharide trans... 91 1e-18
TC87126 similar to GP|15724240|gb|AAL06513.1 At1g75220/F22H5_6 {... 91 1e-18
TC78543 similar to PIR|T14545|T14545 probable sugar transporter ... 90 3e-18
TC89747 similar to PIR|F96696|F96696 protein F1N21.12 [imported]... 88 1e-17
TC83804 similar to GP|9759246|dbj|BAB09770.1 sugar transporter-l... 87 2e-17
TC82331 weakly similar to EGAD|120604|128852 hypothetical protei... 85 9e-17
TC91198 similar to GP|16648753|gb|AAL25568.1 AT5g16150/T21H19_70... 84 2e-16
>TC77798 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_140
{Arabidopsis thaliana}, partial (88%)
Length = 2467
Score = 844 bits (2180), Expect = 0.0
Identities = 435/596 (72%), Positives = 504/596 (83%), Gaps = 4/596 (0%)
Frame = +3
Query: 1 MSGAVIVAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPTVEGLIVAMSLIGATVVT 60
M GAV+VA+AA+IGN LQGWDNATIAGSILYIK++ LQ+ T+EGL+VAMSLIGATV+T
Sbjct: 111 MKGAVLVAIAASIGNFLQGWDNATIAGSILYIKKDLALQT--TMEGLVVAMSLIGATVIT 284
Query: 61 TCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLY 120
TCSG +SD GRRPM+IISS+LYFL SLVM WSPNVY+L ARLLDG GIGLAVTLVP+Y
Sbjct: 285 TCSGPISDWLGRRPMMIISSVLYFLGSLVMLWSPNVYVLCLARLLDGFGIGLAVTLVPVY 464
Query: 121 ISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPSWRLMLGVLSIPSLIYFAL 180
ISE AP +IRGSLNTLPQF+GS GMF SYCMVF MSL+ +PSWR+MLGVLSIPSL YF L
Sbjct: 465 ISETAPSDIRGSLNTLPQFSGSGGMFLSYCMVFVMSLSPSPSWRIMLGVLSIPSLFYFLL 644
Query: 181 TLLLLPESPRWLVSKGRMLEAKKVLQRLRGCQDVAGEMALLVEGLGVGGDTSIEEYIIGP 240
T+ LPESPRWLVSKG+MLEAKKVLQRLRG DV+GEMALLVEGLG+GGD SIEEYIIGP
Sbjct: 645 TVFFLPESPRWLVSKGKMLEAKKVLQRLRGQDDVSGEMALLVEGLGIGGDASIEEYIIGP 824
Query: 241 DNELADEEDPSTGKDQIKLYGPEHGQSWVARPVTGQSSVGLVSRKGSMANPSGLVDPLVT 300
+E+ D + +T KD+I+LYG + G SW+A+PVTGQSS+GLVSR GS+ ++DPLVT
Sbjct: 825 ADEVGDGHEQTTDKDKIRLYGSQAGLSWLAKPVTGQSSLGLVSRHGSL-----VMDPLVT 989
Query: 301 LFGSVHEKLPETGSMRSTLFPHFGSMFSVGGNQPRNEDWDEESLAREGDDYISDAAAGDS 360
LFGS+HEKLPETGSMRS LFP+FGSMFS + E WDEESL REG+DY+SD AAGD+
Sbjct: 990 LFGSIHEKLPETGSMRSALFPNFGSMFSTAEPHIKTEHWDEESLQREGEDYVSDGAAGDT 1169
Query: 361 DDNLQSPLISRQTTSMDKDM-PLPAQGSLSN-MRQGSLLQGNAGEPVGSTGIGGGWQLAW 418
DD+L SPLISRQTTS++KD+ P P+ GSL N MR+ S L +GEPVGSTGIGGGWQLAW
Sbjct: 1170DDDLHSPLISRQTTSLEKDLPPPPSHGSLLNSMRRHSSLMQESGEPVGSTGIGGGWQLAW 1349
Query: 419 KWSEQEGPGGKKEGGFKRIYLHQE--GGPGSIRASVVSLPGGDVPTDGDVVQQAAALVSQ 476
KWS +G GKK+G FKRIYLH+E G GS R S+VS+PG +GD V QAAALVSQ
Sbjct: 1350KWS-GKGEDGKKQGEFKRIYLHEEGVGVSGSRRGSMVSIPG-----EGDFV-QAAALVSQ 1508
Query: 477 PALYNKELMHQQPVGPAMIHPSETAAKGPSWNDLFEPGVKHALFVGVGLQILQQFSGING 536
PALY+KEL+ +QPVGPAMIHPS+TA+KGP W L EPGVKHAL VG+G+Q+LQQFSGING
Sbjct: 1509PALYSKELIGEQPVGPAMIHPSKTASKGPIWEALLEPGVKHALIVGIGIQLLQQFSGING 1688
Query: 537 VLYYTPQILEQAGVGYLLSNLGLSSTSSSFLISAVTTLLMLPCIAVAMRLMDISGR 592
VLYYTPQILE+AGV LL++LGLSSTSSSFLISAVTTLLMLP I +AMRLMD++GR
Sbjct: 1689VLYYTPQILEEAGVAVLLADLGLSSTSSSFLISAVTTLLMLPSIGLAMRLMDVTGR 1856
>TC79692 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_140
{Arabidopsis thaliana}, partial (49%)
Length = 1353
Score = 392 bits (1008), Expect(3) = 0.0
Identities = 202/203 (99%), Positives = 203/203 (99%)
Frame = +2
Query: 1 MSGAVIVAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPTVEGLIVAMSLIGATVVT 60
MSGAVIVAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPTVEGLIVAMSLIGATVVT
Sbjct: 152 MSGAVIVAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPTVEGLIVAMSLIGATVVT 331
Query: 61 TCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLY 120
TCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLY
Sbjct: 332 TCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLY 511
Query: 121 ISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPSWRLMLGVLSIPSLIYFAL 180
ISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPSWRLMLGVLSIPSLIYFAL
Sbjct: 512 ISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPSWRLMLGVLSIPSLIYFAL 691
Query: 181 TLLLLPESPRWLVSKGRMLEAKK 203
TLL+LPESPRWLVSKGRMLEAKK
Sbjct: 692 TLLVLPESPRWLVSKGRMLEAKK 760
Score = 380 bits (976), Expect(3) = 0.0
Identities = 188/194 (96%), Positives = 189/194 (96%)
Frame = +3
Query: 195 KGRMLEAKKVLQRLRGCQDVAGEMALLVEGLGVGGDTSIEEYIIGPDNELADEEDPSTGK 254
K L +KVLQRLRGCQDVAGEMALLVEGLGVGGDTSIEEYIIGPDNELADEEDPSTGK
Sbjct: 735 KAECLRLRKVLQRLRGCQDVAGEMALLVEGLGVGGDTSIEEYIIGPDNELADEEDPSTGK 914
Query: 255 DQIKLYGPEHGQSWVARPVTGQSSVGLVSRKGSMANPSGLVDPLVTLFGSVHEKLPETGS 314
DQIKLYGPEHGQSWVARPVTGQSSVGLVSRKGSMANPSGLVDPLVTLFGSVHEKLPETGS
Sbjct: 915 DQIKLYGPEHGQSWVARPVTGQSSVGLVSRKGSMANPSGLVDPLVTLFGSVHEKLPETGS 1094
Query: 315 MRSTLFPHFGSMFSVGGNQPRNEDWDEESLAREGDDYISDAAAGDSDDNLQSPLISRQTT 374
MRSTLFPHFGSMFSVGGNQPRNEDWDEESLAREGDDYISDAAAGDSDDNLQSPLISRQTT
Sbjct: 1095MRSTLFPHFGSMFSVGGNQPRNEDWDEESLAREGDDYISDAAAGDSDDNLQSPLISRQTT 1274
Query: 375 SMDKDMPLPAQGSL 388
SMDKDMPLPAQGSL
Sbjct: 1275SMDKDMPLPAQGSL 1316
Score = 28.1 bits (61), Expect(3) = 0.0
Identities = 12/14 (85%), Positives = 13/14 (92%)
Frame = +1
Query: 387 SLSNMRQGSLLQGN 400
+ SNMRQGSLLQGN
Sbjct: 1312 AFSNMRQGSLLQGN 1353
>TC87914 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_140
{Arabidopsis thaliana}, partial (39%)
Length = 1129
Score = 338 bits (866), Expect(2) = e-104
Identities = 169/169 (100%), Positives = 169/169 (100%)
Frame = +3
Query: 424 EGPGGKKEGGFKRIYLHQEGGPGSIRASVVSLPGGDVPTDGDVVQQAAALVSQPALYNKE 483
EGPGGKKEGGFKRIYLHQEGGPGSIRASVVSLPGGDVPTDGDVVQQAAALVSQPALYNKE
Sbjct: 3 EGPGGKKEGGFKRIYLHQEGGPGSIRASVVSLPGGDVPTDGDVVQQAAALVSQPALYNKE 182
Query: 484 LMHQQPVGPAMIHPSETAAKGPSWNDLFEPGVKHALFVGVGLQILQQFSGINGVLYYTPQ 543
LMHQQPVGPAMIHPSETAAKGPSWNDLFEPGVKHALFVGVGLQILQQFSGINGVLYYTPQ
Sbjct: 183 LMHQQPVGPAMIHPSETAAKGPSWNDLFEPGVKHALFVGVGLQILQQFSGINGVLYYTPQ 362
Query: 544 ILEQAGVGYLLSNLGLSSTSSSFLISAVTTLLMLPCIAVAMRLMDISGR 592
ILEQAGVGYLLSNLGLSSTSSSFLISAVTTLLMLPCIAVAMRLMDISGR
Sbjct: 363 ILEQAGVGYLLSNLGLSSTSSSFLISAVTTLLMLPCIAVAMRLMDISGR 509
Score = 59.3 bits (142), Expect(2) = e-104
Identities = 24/24 (100%), Positives = 24/24 (100%)
Frame = +2
Query: 593 SISFHISPREPCGFGRHCKCVYLN 616
SISFHISPREPCGFGRHCKCVYLN
Sbjct: 545 SISFHISPREPCGFGRHCKCVYLN 616
>TC84406 similar to GP|12039327|gb|AAG46115.1 putative sugar transporter
{Oryza sativa}, partial (13%)
Length = 753
Score = 185 bits (469), Expect = 5e-47
Identities = 103/199 (51%), Positives = 129/199 (64%), Gaps = 4/199 (2%)
Frame = +3
Query: 395 SLLQGNAGEPVGSTGIGGGWQLAWKWSEQEGPGGKKEGGFKRIYLHQEGGPGSI----RA 450
SL+ GN +T IGGGWQL +K S + GGK+EG +R+YLH + ++
Sbjct: 117 SLVHGNYVGTPRNTDIGGGWQLVYK-STDDAMGGKREG-LQRVYLHADTSAAAVSQSPHV 290
Query: 451 SVVSLPGGDVPTDGDVVQQAAALVSQPALYNKELMHQQPVGPAMIHPSETAAKGPSWNDL 510
S VS G D+P DG QAA +VS+ L + + ETAAKGP W L
Sbjct: 291 SFVSTSGYDIPIDGGEAFQAAGIVSRSILGTSDALSVP----------ETAAKGPKWRAL 440
Query: 511 FEPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGVGYLLSNLGLSSTSSSFLISA 570
EPGVK AL VG+GLQILQQ +GING LYY PQILEQAGVG LLSNLG+SS S+SFL++
Sbjct: 441 LEPGVKRALIVGIGLQILQQAAGINGFLYYAPQILEQAGVGALLSNLGISSISASFLVNI 620
Query: 571 VTTLLMLPCIAVAMRLMDI 589
+TT MLPCIA+A++LMD+
Sbjct: 621 ITTFCMLPCIAIAIKLMDV 677
>TC78321 similar to GP|21689841|gb|AAM67564.1 unknown protein {Arabidopsis
thaliana}, partial (80%)
Length = 2093
Score = 154 bits (390), Expect = 7e-38
Identities = 86/221 (38%), Positives = 136/221 (60%), Gaps = 2/221 (0%)
Frame = +2
Query: 7 VAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPT-VEGLIVAMSLIGATVVTTCSGA 65
+A +A IG LL G+D I+G++LYI+ EF + T ++ IV+ ++ GA + G
Sbjct: 230 LAFSAGIGGLLFGYDTGVISGALLYIRDEFPAVEKKTWLQEAIVSTAIAGAIIGAAIGGW 409
Query: 66 LSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIA 125
++D FGR+ +I++ L+ L S+++ +PN L+ R+ GLG+G+A PLYISE +
Sbjct: 410 INDRFGRKVSIIVADTLFLLGSIILAAAPNPATLIVGRVFVGLGVGMASMASPLYISEAS 589
Query: 126 PPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAP-SWRLMLGVLSIPSLIYFALTLLL 184
P +RG+L +L F + G F SY + ++ TKAP +WR MLGV + P++I L +L
Sbjct: 590 PTRVRGALVSLNSFLITGGQFLSY--LINLAFTKAPGTWRWMLGVAAAPAVIQIVL-MLS 760
Query: 185 LPESPRWLVSKGRMLEAKKVLQRLRGCQDVAGEMALLVEGL 225
LPESPRWL KG+ EAK +L+++ +D E+ L E +
Sbjct: 761 LPESPRWLYRKGKEEEAKVILKKIYEVEDYDNEIQALKESV 883
Score = 48.1 bits (113), Expect = 1e-05
Identities = 27/78 (34%), Positives = 45/78 (57%)
Frame = +2
Query: 515 VKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGVGYLLSNLGLSSTSSSFLISAVTTL 574
V+ L+ GVGL QQF+GIN V+YY+P I++ A G +S ++ L+S +T+
Sbjct: 944 VRRGLYAGVGLAFFQQFTGINTVMYYSPSIVQLA---------GFASKRTALLLSLITSG 1096
Query: 575 LMLPCIAVAMRLMDISGR 592
L +++ +D +GR
Sbjct: 1097 LNAFGSILSIYFIDKTGR 1150
>TC79361 similar to GP|17380890|gb|AAL36257.1 putative membrane transporter
protein {Arabidopsis thaliana}, partial (55%)
Length = 949
Score = 151 bits (381), Expect = 8e-37
Identities = 84/217 (38%), Positives = 134/217 (61%), Gaps = 2/217 (0%)
Frame = +1
Query: 7 VAVAAAIGNLLQGWDNATIAGSILYIKREF-QLQSEPTVEGLIVAMSLIGATVVTTCSGA 65
+A A IG LL G+D I+G++LYIK +F Q+++ ++ IV+M++ GA V G
Sbjct: 178 LAAVAGIGGLLFGYDTGVISGALLYIKDDFPQVRNSNFLQETIVSMAIAGAIVGAAFGGW 357
Query: 66 LSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIA 125
L+D +GR+ +++ +++ L +++M +P+ Y+L+ RLL GLG+G+A P+YI+E+A
Sbjct: 358 LNDAYGRKKATLLADVIFILGAILMAAAPDPYVLIAGRLLVGLGVGIASVTAPVYIAEVA 537
Query: 126 PPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAP-SWRLMLGVLSIPSLIYFALTLLL 184
P EIRGSL + + G F SY + + T+ P +WR MLGV +P+LI F + +L
Sbjct: 538 PSEIRGSLVSTNVLMITGGQFVSY--LVNLVFTQVPGTWRWMLGVSGVPALIQF-ICMLF 708
Query: 185 LPESPRWLVSKGRMLEAKKVLQRLRGCQDVAGEMALL 221
LPESPRWL K R EA V+ ++ + E+ L
Sbjct: 709 LPESPRWLFIKNRKNEAVDVISKIYDLSRLEDEIDFL 819
>TC77077 similar to GP|9293909|dbj|BAB01812.1 sugar transporter protein
{Arabidopsis thaliana}, partial (85%)
Length = 1752
Score = 136 bits (342), Expect = 3e-32
Identities = 81/232 (34%), Positives = 141/232 (59%), Gaps = 5/232 (2%)
Frame = +1
Query: 4 AVIVAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPTVE---GLIVAMSLIGATVVT 60
A A+ A++ ++L G+D ++G+ +YIKR+ ++ S+ +E G+I SLIG+ +
Sbjct: 109 AFACAMLASMTSILLGYDIGVMSGAAIYIKRDLKV-SDGKIEVLLGIINIYSLIGSCL-- 279
Query: 61 TCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLY 120
+G SD GRR ++ + ++F+ +L+M +SPN L+F R + G+GIG A+ + P+Y
Sbjct: 280 --AGRTSDWIGRRYTIVFAGAIFFVGALLMGFSPNYNFLMFGRFVAGVGIGYALMIAPVY 453
Query: 121 ISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMS-LTKAPSWRLMLGVLSIPSLIYFA 179
+E++P RG L + P+ ++G+ Y + S L+ WR+MLGV +IPS+I A
Sbjct: 454 TAEVSPASSRGFLTSFPEVFINSGILLGYISNYAFSKLSLKLGWRMMLGVGAIPSVI-LA 630
Query: 180 LTLLLLPESPRWLVSKGRMLEAKKVLQRLRGCQDVAG-EMALLVEGLGVGGD 230
+ +L +PESPRWLV +GR+ +A KVL + ++ A +A + + G+ D
Sbjct: 631 VGVLAMPESPRWLVMRGRLGDAIKVLNKTSDSKEEAQLRLAEIKQAAGIPED 786
Score = 43.5 bits (101), Expect = 2e-04
Identities = 21/75 (28%), Positives = 40/75 (53%), Gaps = 7/75 (9%)
Frame = +1
Query: 507 WNDLF---EPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGVG----YLLSNLGL 559
W +LF P V+H + +G+ QQ SG++ V+ Y+P I ++AG+ L++ + +
Sbjct: 838 WKELFLYPTPAVRHIVIAALGIHFFQQASGVDAVVLYSPTIFKKAGINGDTHLLIATIAV 1017
Query: 560 SSTSSSFLISAVTTL 574
+ F++ A L
Sbjct: 1018GFVKTLFILVATFML 1062
>TC89865 similar to GP|17380890|gb|AAL36257.1 putative membrane transporter
protein {Arabidopsis thaliana}, partial (33%)
Length = 617
Score = 109 bits (273), Expect = 3e-24
Identities = 56/144 (38%), Positives = 92/144 (63%), Gaps = 1/144 (0%)
Frame = +1
Query: 11 AAIGNLLQGWDNATIAGSILYIKREFQ-LQSEPTVEGLIVAMSLIGATVVTTCSGALSDL 69
A+IG LL G+D I+G++LYIK +FQ ++ ++ IV+M++ GA V G ++D
Sbjct: 112 ASIGGLLFGYDTGVISGALLYIKDDFQAVRYSHFLQETIVSMAVAGAIVGAAVGGWMNDR 291
Query: 70 FGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIAPPEI 129
+GR+ II+ +++ L ++VM +P+ YIL+ R+L GLG+G+A P+YI+E++P EI
Sbjct: 292 YGRKKATIIADVIFILGAIVMAAAPDPYILILGRVLVGLGVGIASVTAPVYIAELSPSEI 471
Query: 130 RGSLNTLPQFAGSAGMFFSYCMVF 153
RG L + G F SY ++F
Sbjct: 472 RGGLVATNVLMITGGQFISYLVIF 543
>TC81325 similar to GP|17380890|gb|AAL36257.1 putative membrane transporter
protein {Arabidopsis thaliana}, partial (32%)
Length = 636
Score = 105 bits (261), Expect = 7e-23
Identities = 52/128 (40%), Positives = 85/128 (65%), Gaps = 1/128 (0%)
Frame = +3
Query: 7 VAVAAAIGNLLQGWDNATIAGSILYIKREFQ-LQSEPTVEGLIVAMSLIGATVVTTCSGA 65
V AA IG LL G+D I+G++LYIK +F +++ ++ IV+M+L+GA + G
Sbjct: 177 VTAAAGIGGLLFGYDTGVISGALLYIKDDFDDVRNSSFLQETIVSMALVGAIIGAATGGW 356
Query: 66 LSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIA 125
++D FGR+ + + +++ L S+VM +P+ Y+L+ RLL G+G+G+A P+YI+E +
Sbjct: 357 INDAFGRKKATLSADVVFTLGSVVMASAPDAYVLILGRLLVGIGVGVASVTAPVYIAESS 536
Query: 126 PPEIRGSL 133
P EIRGSL
Sbjct: 537 PSEIRGSL 560
>TC84386 similar to GP|21689841|gb|AAM67564.1 unknown protein {Arabidopsis
thaliana}, partial (30%)
Length = 664
Score = 104 bits (259), Expect = 1e-22
Identities = 57/156 (36%), Positives = 94/156 (59%), Gaps = 1/156 (0%)
Frame = +2
Query: 7 VAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPT-VEGLIVAMSLIGATVVTTCSGA 65
+A +A IG L G+D I+G++LYI+ +F+ T ++ IV+ +L GA + + G
Sbjct: 194 LAFSAGIGGFLFGYDTGVISGALLYIRDDFKAVDRQTWLQEAIVSTALAGAIIGASVGGW 373
Query: 66 LSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIA 125
++D FGR+ +I++ L+F+ S++M + N IL+ R+ GLG+G+A PLYISE +
Sbjct: 374 INDRFGRKKAIILADALFFIGSVIMAAAINPAILIVGRVFVGLGVGMASMASPLYISEAS 553
Query: 126 PPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAP 161
P +RG+L +L F + G SY V ++ T AP
Sbjct: 554 PTRVRGALVSLNGFLITGGQXLSY--VINLAFTNAP 655
>TC87421 sugar transporter
Length = 1753
Score = 93.2 bits (230), Expect = 3e-19
Identities = 67/237 (28%), Positives = 122/237 (51%), Gaps = 20/237 (8%)
Frame = +2
Query: 6 IVAVAAAIGNLLQGWDNATIAGSIL----YIKREFQL---------------QSEPTVEG 46
I + AA+G L+ G+D I+G + ++K+ F Q +
Sbjct: 101 ITCIVAAMGGLIFGYDIG-ISGGVTSMDPFLKKFFPAVYRKKNKDKSTNQYCQYDSQTLT 277
Query: 47 LIVAMSLIGATVVTTCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLD 106
+ + + A + + + ++ FGR+ ++ LL+ + +L+ ++ +V++L+ R+L
Sbjct: 278 MFTSSLYLAALLSSLVASTITRRFGRKLSMLFGGLLFLVGALINGFANHVWMLIVGRILL 457
Query: 107 GLGIGLAVTLVPLYISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAP-SWRL 165
G GIG A VPLY+SE+AP + RG+LN Q + + G+ + + + + K WRL
Sbjct: 458 GFGIGFANQPVPLYLSEMAPYKYRGALNIGFQLSITIGILVANVLNYFFAKIKGGWGWRL 637
Query: 166 MLGVLSIPSLIYFALTLLLLPESPRWLVSKGRMLEAKKVLQRLRGCQDVAGEMALLV 222
LG +P+LI + L+LP++P ++ +G AK L+R+RG +DV E LV
Sbjct: 638 SLGGAMVPALI-ITIGSLVLPDTPNSMIERGDRDGAKAQLKRIRGIEDVDEEFNDLV 805
Score = 33.1 bits (74), Expect = 0.32
Identities = 20/90 (22%), Positives = 40/90 (44%)
Frame = +2
Query: 507 WNDLFEPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGVGYLLSNLGLSSTSSSF 566
W +L + + L + V + QQF+GIN +++Y P + G +S
Sbjct: 842 WRNLLQRKYRPQLTMAVLIPFFQQFTGINVIMFYAPVLFNSIGF----------KDDASL 991
Query: 567 LISAVTTLLMLPCIAVAMRLMDISGRSISF 596
+ + +T ++ + V++ +D GR F
Sbjct: 992 MSAVITGVVNVVATCVSIYGVDKWGRRALF 1081
>BQ751130 weakly similar to GP|15289796|dbj putative monosaccharide
transporter 3 {Oryza sativa (japonica cultivar-group)},
partial (7%)
Length = 795
Score = 92.0 bits (227), Expect = 6e-19
Identities = 70/223 (31%), Positives = 109/223 (48%), Gaps = 16/223 (7%)
Frame = +3
Query: 4 AVIVAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPTVEG-------LIVAMSLIGA 56
A ++ A+ G + G+D+ I G +L + + P E LIV++ G
Sbjct: 39 AYVICAFASFGGIFFGYDSGYING-VLGANQFIEHVEGPGAEAISESHQSLIVSILSCGT 215
Query: 57 TVVTTCSGALSDLFGRRPMLIISSLLYFLSSLVMFWS---PNVYILLFARLLDGLGIGLA 113
+G +SD GR+ +II +Y + L+ ++ + ++ RL+ G+G+G
Sbjct: 216 FFGALIAGDVSDWIGRKWTVIIGCGIYMIGVLIQMFTGMGAPLGAIVAGRLIAGIGVGFE 395
Query: 114 VTLVPLYISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPSWRLMLGVLSIP 173
+V LY+SEI P ++RG+L QF + G+ + C+V+ A R GV IP
Sbjct: 396 SAIVILYMSEICPKKVRGALVAGYQFCITIGLLLAACVVY------ATKDRGDTGVYRIP 557
Query: 174 SLIYFALT------LLLLPESPRWLVSKGRMLEAKKVLQRLRG 210
I F LLLLPESPR+ V KGR+ +A L RLRG
Sbjct: 558 IAIQFPWALILGGGLLLLPESPRFFVKKGRIADATAALSRLRG 686
>TC78964 weakly similar to PIR|T00450|T00450 probable monosaccharide
transport protein T14N5.7 - Arabidopsis thaliana,
partial (98%)
Length = 1771
Score = 92.0 bits (227), Expect = 6e-19
Identities = 54/171 (31%), Positives = 94/171 (54%)
Frame = +1
Query: 47 LIVAMSLIGATVVTTCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLD 106
L + A V+T + L+ GR+ +I+ +L + + +++ + N+ L+ R+
Sbjct: 289 LFTSSLYFSALVMTFFASYLTRNKGRKATIIVGALSFLIGAILNAAAQNIPTLIIGRVFL 468
Query: 107 GLGIGLAVTLVPLYISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPSWRLM 166
G GIG VPLY+SE+AP RG++N L QF AG+ + + + WR+
Sbjct: 469 GGGIGFGNQAVPLYLSEMAPASSRGAVNQLFQFTTCAGILIANLVNYFTDKIHPHGWRIS 648
Query: 167 LGVLSIPSLIYFALTLLLLPESPRWLVSKGRMLEAKKVLQRLRGCQDVAGE 217
LG+ IP+++ L + E+P LV +GR+ EA+KVL+++RG ++V E
Sbjct: 649 LGLAGIPAVL-MLLGGIFCAETPNSLVEQGRLDEARKVLEKVRGTKNVDAE 798
>TC86673 weakly similar to PIR|S25015|S25015 monosaccharide transport
protein MST1 - common tobacco, partial (65%)
Length = 1162
Score = 91.3 bits (225), Expect = 1e-18
Identities = 64/219 (29%), Positives = 110/219 (50%), Gaps = 20/219 (9%)
Frame = +2
Query: 5 VIVAVAAAIGNLLQGWDNATIAGSIL---YIKREFQLQSEPT----------------VE 45
+I + AA G L+ G+D+ G ++KR F E +
Sbjct: 98 IITCIMAATGGLIFGYDHGVSGGVTSMDSFLKRFFPSVYEQESNLKPSANQYCKFNSQIL 277
Query: 46 GLIVAMSLIGATVVTTCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLL 105
L + I A V + +++ GRR +I+ + + +L+ + N+ +L+ RLL
Sbjct: 278 TLFTSSLYISALVAGLGASSITRALGRRTTMILGGIFFVSGALLNGLAMNIAMLIAGRLL 457
Query: 106 DGLGIGLAVTLVPLYISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMS-LTKAPSWR 164
G GIG A VP+Y+SE+AP + RG+LN Q + + G+F + + S + WR
Sbjct: 458 LGFGIGCANQAVPIYLSEMAPYKYRGALNMCFQLSITIGIFTANMFNYYFSKILNGEGWR 637
Query: 165 LMLGVLSIPSLIYFALTLLLLPESPRWLVSKGRMLEAKK 203
L LG+ ++P+++ F + + LP+SP LV++GR EA+K
Sbjct: 638 LSLGLGAVPAVV-FIIGSICLPDSPNSLVTRGRHEEARK 751
Score = 33.1 bits (74), Expect = 0.32
Identities = 20/85 (23%), Positives = 39/85 (45%), Gaps = 1/85 (1%)
Frame = +3
Query: 494 MIHPSETAAKGPS-WNDLFEPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGVGY 552
++ SE +AK W L E + L + + QQF+G+N + +Y P + G G
Sbjct: 804 IVAASEASAKVKHPWKTLQERKYRPQLVFAILIPFFQQFTGLNVITFYAPILFRTIGFGS 983
Query: 553 LLSNLGLSSTSSSFLISAVTTLLML 577
S + + S +S + ++ ++
Sbjct: 984 QASLMSAAIIGSFKPVSTLVSIFVV 1058
>TC87126 similar to GP|15724240|gb|AAL06513.1 At1g75220/F22H5_6 {Arabidopsis
thaliana}, partial (79%)
Length = 1524
Score = 91.3 bits (225), Expect = 1e-18
Identities = 55/156 (35%), Positives = 85/156 (54%)
Frame = +3
Query: 55 GATVVTTCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAV 114
GA V SG +++ GR+ L+I+S+ + L + ++ + L RLL+G G+G+
Sbjct: 6 GAMVGAIASGQIAEYVGRKGSLMIASIPNIIGWLAISFAKDSSFLFMGRLLEGFGVGIIS 185
Query: 115 TLVPLYISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPSWRLMLGVLSIPS 174
+VP+YI+EIAP +RGSL ++ Q + + G+ +Y L +WR +L +L I
Sbjct: 186 YVVPVYIAEIAPENMRGSLGSVNQLSVTIGIMLAYL------LGLFANWR-VLAILGILP 344
Query: 175 LIYFALTLLLLPESPRWLVSKGRMLEAKKVLQRLRG 210
L +PESPRWL G M E + LQ LRG
Sbjct: 345 CTVLIPGLFFIPESPRWLAKMGMMEEFETSLQVLRG 452
Score = 48.1 bits (113), Expect = 1e-05
Identities = 31/81 (38%), Positives = 44/81 (54%)
Frame = +3
Query: 519 LFVGVGLQILQQFSGINGVLYYTPQILEQAGVGYLLSNLGLSSTSSSFLISAVTTLLMLP 578
L VG+GL +LQQ SGINGVL+Y+ I AG+ SS +++ + A+ +
Sbjct: 567 LSVGIGLLVLQQLSGINGVLFYSTSIFANAGIS--------SSNAATVGLGAIQVI---- 710
Query: 579 CIAVAMRLMDISGRSISFHIS 599
VA L+D SGR + IS
Sbjct: 711 ATGVATWLVDKSGRRVLLIIS 773
Score = 28.5 bits (62), Expect = 7.9
Identities = 43/161 (26%), Positives = 71/161 (43%), Gaps = 18/161 (11%)
Frame = +3
Query: 58 VVTTCSGALSDLFGRRPMLIISSLLYFLSSLVM---------------FWSPNVYILLFA 102
+ T + L D GRR +LIISS L S LV+ ++S I +
Sbjct: 708 IATGVATWLVDKSGRRVLLIISSSLMTASLLVVSIAFYLEGVVEKDSQYFSILGIISVVG 887
Query: 103 RLLDGLGIGLAVTLVP-LYISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAP 161
++ +G L + +P L +SEI P I+G AGS ++ + + +++T
Sbjct: 888 LVVMVIGFSLGLGPIPWLIMSEILPVNIKG-------LAGSTATMANWLVAWIITMTA-- 1040
Query: 162 SWRLMLGVLSIPS-LIYFALTLLLLPESPRWL-VSKGRMLE 200
L+L S + LIY + + + W+ +KGR LE
Sbjct: 1041--NLLLTWSSGGTFLIYTVVAAFTVVFTSLWVPETKGRTLE 1157
>TC78543 similar to PIR|T14545|T14545 probable sugar transporter protein -
beet, partial (93%)
Length = 1653
Score = 89.7 bits (221), Expect = 3e-18
Identities = 57/173 (32%), Positives = 96/173 (54%), Gaps = 1/173 (0%)
Frame = +1
Query: 47 LIVAMSLIGATVVTTCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLD 106
L ++S +GA V SG +++ GR+ L+I+S+ + L++ ++ + L RLL+
Sbjct: 271 LFGSLSNVGAMVGAIASGQIAEYMGRKGSLMIASIPNIIGWLMISFANDSSFLYMGRLLE 450
Query: 107 GLGIGLAVTLVPLYISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPSWRLM 166
G G+G+ VP+YI+EI+P +RGSL ++ Q + + G+ +Y + G+ + WR
Sbjct: 451 GFGVGIISYTVPVYIAEISPQNLRGSLVSVNQLSVTLGIMLAY--LLGLFV----EWR-F 609
Query: 167 LGVLSIPSLIYFALTLLLLPESPRWLVSKGRMLEAKKVLQRLRGCQ-DVAGEM 218
L +L I L +PESPRWL G E + LQ LRG + D++ E+
Sbjct: 610 LAILGIIPCTLLIPGLFFIPESPRWLAKMGMTEEFENSLQVLRGFETDISVEV 768
Score = 48.9 bits (115), Expect = 6e-06
Identities = 30/81 (37%), Positives = 44/81 (54%)
Frame = +1
Query: 519 LFVGVGLQILQQFSGINGVLYYTPQILEQAGVGYLLSNLGLSSTSSSFLISAVTTLLMLP 578
L +G+GL +LQQ SGINGVL+Y+ I + AG+ SS ++F + AV L
Sbjct: 856 LMIGIGLLVLQQLSGINGVLFYSSTIFQNAGIS--------SSDVATFGVGAVQVL---- 999
Query: 579 CIAVAMRLMDISGRSISFHIS 599
+ + L D SGR + +S
Sbjct: 1000ATTLTLWLADKSGRRLLLIVS 1062
>TC89747 similar to PIR|F96696|F96696 protein F1N21.12 [imported] -
Arabidopsis thaliana, partial (64%)
Length = 1372
Score = 87.8 bits (216), Expect = 1e-17
Identities = 61/214 (28%), Positives = 103/214 (47%), Gaps = 1/214 (0%)
Frame = +3
Query: 9 VAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPTVEGLIVAMSLIGATVVTTCSGALSD 68
+ A I + L G+ + + I + EGL+V++ L GA SG ++D
Sbjct: 387 LVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALFGCLLSGWIAD 566
Query: 69 LFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIAPPE 128
GRR + +L + + + + N++ +L RL G G+GL + LY++E++P
Sbjct: 567 AVGRRRAFQLCALPMIIGAAMSAATNNLFGMLVGRLFVGTGLGLGPPVAALYVTEVSPAF 746
Query: 129 IRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPS-WRLMLGVLSIPSLIYFALTLLLLPE 187
+RG+ L Q A G+ S + G+ + + WR+ V +IP+ I AL + E
Sbjct: 747 VRGTYGALIQIATCFGILGS--LFIGIPVKEISGWWRVCFWVSTIPAAI-LALAMDFCAE 917
Query: 188 SPRWLVSKGRMLEAKKVLQRLRGCQDVAGEMALL 221
SP WL +GR EA+ +RL G + M+ L
Sbjct: 918 SPHWLYKQGRTAEAEAEFERLLGVSEAKFAMSQL 1019
Score = 35.4 bits (80), Expect = 0.065
Identities = 18/51 (35%), Positives = 31/51 (60%), Gaps = 2/51 (3%)
Frame = +3
Query: 519 LFVGVGLQILQQFSGINGVLYYTPQILEQAGVGYLLSN--LGLSSTSSSFL 567
+F+G L LQQ SGIN V Y++ + + AGV +N +G+++ + S +
Sbjct: 1095 VFIGSTLFALQQLSGINAVFYFSSTVFKSAGVPSDFANVCIGVANLTGSII 1247
>TC83804 similar to GP|9759246|dbj|BAB09770.1 sugar transporter-like protein
{Arabidopsis thaliana}, partial (29%)
Length = 721
Score = 86.7 bits (213), Expect = 2e-17
Identities = 58/150 (38%), Positives = 80/150 (52%), Gaps = 9/150 (6%)
Frame = +3
Query: 70 FGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIAPPEI 129
F R+ LI ++LLY L S + +P + +LL RLL GLGIGLA+ PLYI+E P +I
Sbjct: 237 FRRKRQLIGAALLYVLGSAITATAPELGVLLAGRLLYGLGIGLAMHGAPLYIAETCPSQI 416
Query: 130 RGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPSWRLMLGVLSIPSLIYFALTLLLLPESP 189
RG+L +L + G+ Y V ++ WR M G S P + L + LPESP
Sbjct: 417 RGTLVSLKELFIVLGILLGY-FVGSFQISTVGGWRFMYG-FSAPLAVLMGLGMWTLPESP 590
Query: 190 RWLV-----SKGRMLEAKK----VLQRLRG 210
RWL+ KG + K+ L +LRG
Sbjct: 591 RWLLLNAVQGKGSFQDLKEKAIVSLGKLRG 680
>TC82331 weakly similar to EGAD|120604|128852 hypothetical protein M01F1.5
{Caenorhabditis elegans}, partial (10%)
Length = 848
Score = 84.7 bits (208), Expect = 9e-17
Identities = 58/204 (28%), Positives = 108/204 (52%), Gaps = 15/204 (7%)
Frame = +3
Query: 4 AVIVAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPT-----------VEGLIVAMS 52
A+++ V +IG L G+D I+ +L+ + + P ++ L+V++
Sbjct: 240 ALVLGVVTSIGGFLFGYDTGQISSMLLFTDFRQRFATGPEDANGVKTWVSIIQSLMVSLM 419
Query: 53 LIGATVVTTCSGALS-DLFGRRPMLIISSLLYFLSSLVMFWSPNVYI-LLFARLLDGLGI 110
IG T++ SGA + D +GRR L +++ + ++V + + ++ ++ R + GLG+
Sbjct: 420 SIG-TLIGALSGAYTADWWGRRKSLSFGVVVFVIGNIVQITAMDTWVHMMMGRFIAGLGV 596
Query: 111 GLAVTLVPLYISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKA--PSWRLMLG 168
G VP++ SE +P EIRG++ Q + G+ S + FG+ + SWR+++G
Sbjct: 597 GNLSVGVPMFQSECSPKEIRGAVVASYQLMITFGILISNLINFGVRNIQESDASWRIVIG 776
Query: 169 VLSIPSLIYFALTLLLLPESPRWL 192
L I + L +L++PESPRWL
Sbjct: 777 -LGIAFAMPLGLGILVVPESPRWL 845
>TC91198 similar to GP|16648753|gb|AAL25568.1 AT5g16150/T21H19_70
{Arabidopsis thaliana}, partial (31%)
Length = 673
Score = 83.6 bits (205), Expect = 2e-16
Identities = 47/133 (35%), Positives = 76/133 (56%), Gaps = 1/133 (0%)
Frame = +2
Query: 2 SGAVIVAVAAA-IGNLLQGWDNATIAGSILYIKREFQLQSEPTVEGLIVAMSLIGATVVT 60
SG V V A +G L G+ + G++ Y+ ++ ++ ++G IV+ L GATV +
Sbjct: 212 SGTVFPYVGVACLGAFLFGYHLGVVNGALEYLAKDLRIAQNTVLQGWIVSTLLAGATVGS 391
Query: 61 TCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLY 120
GAL+D FGR + ++ + + + +V ++ R L G+GIG+A +VPLY
Sbjct: 392 FTGGALADKFGRTRTFQLDAIPLAIGGFLCATAQSVQTMIVGRSLAGIGIGIASAIVPLY 571
Query: 121 ISEIAPPEIRGSL 133
ISEI+P EIRG+L
Sbjct: 572 ISEISPTEIRGAL 610
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.137 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,891,829
Number of Sequences: 36976
Number of extensions: 267283
Number of successful extensions: 1595
Number of sequences better than 10.0: 80
Number of HSP's better than 10.0 without gapping: 1526
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1558
length of query: 616
length of database: 9,014,727
effective HSP length: 102
effective length of query: 514
effective length of database: 5,243,175
effective search space: 2694991950
effective search space used: 2694991950
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC121239.3


