

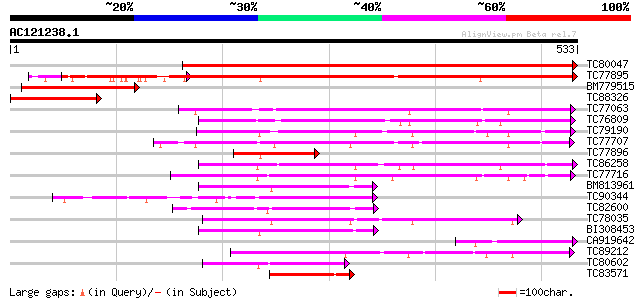
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121238.1 + phase: 0
(533 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC80047 similar to GP|18087577|gb|AAL58919.1 AT4g00150/F6N15_20 ... 745 0.0
TC77895 similar to GP|18087577|gb|AAL58919.1 AT4g00150/F6N15_20 ... 517 e-147
BM779515 weakly similar to GP|18087577|gb| AT4g00150/F6N15_20 {A... 234 6e-62
TC88326 similar to GP|18087577|gb|AAL58919.1 AT4g00150/F6N15_20 ... 152 4e-37
TC77063 similar to GP|20334379|gb|AAM19210.1 GAI-like protein 1 ... 130 1e-30
TC76809 similar to GP|20334379|gb|AAM19210.1 GAI-like protein 1 ... 123 2e-28
TC79190 similar to PIR|E96542|E96542 scarecrow-like protein [imp... 122 4e-28
TC77707 weakly similar to GP|8777405|dbj|BAA96995.1 SCARECROW ge... 110 2e-24
TC77896 weakly similar to GP|22087131|gb|AAM90848.1 hairy merist... 110 2e-24
TC86258 similar to GP|16930433|gb|AAL31902.1 AT4g17230/dl4650c {... 109 3e-24
TC77716 similar to GP|20465587|gb|AAM20276.1 unknown protein {Ar... 106 3e-23
BM813961 homologue to GP|13365610|db SCARECROW {Pisum sativum}, ... 89 4e-18
TC90344 weakly similar to PIR|H86282|H86282 protein F10B6.34 [im... 87 1e-17
TC82600 similar to GP|9757937|dbj|BAB08425.1 SCARECROW gene regu... 84 1e-16
TC78035 similar to GP|8778540|gb|AAF79548.1| F22G5.9 {Arabidopsi... 83 3e-16
BI308453 weakly similar to PIR|T02736|T0 probable SCARECROW gene... 81 9e-16
CA919642 similar to PIR|E85433|E854 SCARECROW-like protein [impo... 78 7e-15
TC89212 weakly similar to PIR|T02531|T02531 probable SCARECROW g... 71 1e-12
TC80602 similar to GP|14517552|gb|AAK62666.1 F17J6.12/F17J6.12 {... 70 2e-12
TC83571 similar to PIR|T10552|T10552 hypothetical protein T12G13... 68 8e-12
>TC80047 similar to GP|18087577|gb|AAL58919.1 AT4g00150/F6N15_20
{Arabidopsis thaliana}, partial (30%)
Length = 1357
Score = 745 bits (1924), Expect = 0.0
Identities = 370/371 (99%), Positives = 371/371 (99%)
Frame = +2
Query: 163 GDDASIQLQQSIFDQLFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRASFYMKEAL 222
G+DASIQLQQSIFDQLFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRASFYMKEAL
Sbjct: 5 GNDASIQLQQSIFDQLFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRASFYMKEAL 184
Query: 223 QLMLHSNGNNLTAFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVI 282
QLMLHSNGNNLTAFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVI
Sbjct: 185 QLMLHSNGNNLTAFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVI 364
Query: 283 DFDIGFGVQWSSFMQEIVLRSNGKPSLKITAVVSPSSCNEIELNFTQENLSQYAKDLNIL 342
DFDIGFGVQWSSFMQEIVLRSNGKPSLKITAVVSPSSCNEIELNFTQENLSQYAKDLNIL
Sbjct: 365 DFDIGFGVQWSSFMQEIVLRSNGKPSLKITAVVSPSSCNEIELNFTQENLSQYAKDLNIL 544
Query: 343 FEFNVLNIESLNLPSCPLPGHFFDSNEAIGVNFPVSSFISNPSCFPVALHFLKQLRPKIV 402
FEFNVLNIESLNLPSCPLPGHFFDSNEAIGVNFPVSSFISNPSCFPVALHFLKQLRPKIV
Sbjct: 545 FEFNVLNIESLNLPSCPLPGHFFDSNEAIGVNFPVSSFISNPSCFPVALHFLKQLRPKIV 724
Query: 403 VTLDKNCDRMDVPLPTNVVHVLQCYSALLESLDAVNVNLDVLQKIERHYIQPTINKIVLS 462
VTLDKNCDRMDVPLPTNVVHVLQCYSALLESLDAVNVNLDVLQKIERHYIQPTINKIVLS
Sbjct: 725 VTLDKNCDRMDVPLPTNVVHVLQCYSALLESLDAVNVNLDVLQKIERHYIQPTINKIVLS 904
Query: 463 HHNQRDKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQR 522
HHNQRDKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQR
Sbjct: 905 HHNQRDKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQR 1084
Query: 523 KELISVSTWRC 533
KELISVSTWRC
Sbjct: 1085KELISVSTWRC 1117
>TC77895 similar to GP|18087577|gb|AAL58919.1 AT4g00150/F6N15_20 {Arabidopsis
thaliana}, partial (41%)
Length = 2591
Score = 517 bits (1331), Expect = e-147
Identities = 290/533 (54%), Positives = 354/533 (66%), Gaps = 48/533 (9%)
Frame = +3
Query: 49 SAGLNKILQNSGYGSQNVDFHGGFGVLDHQQQQGLTMMDASVQQG---NYNVFPFIPE-- 103
S G+N+I QN + S V G G+ QQQ + +D + N N F+P
Sbjct: 840 SRGMNQI-QNPNFSSSQVSISHG-GLFHQHQQQSIGSLDQKLPHQFVMNQNQAQFMPNPS 1013
Query: 104 -----------------------------NYNV--LPLLDSGQEVFARR---HQQQE--T 127
NY + +P LDSGQE+F RR HQQQ+
Sbjct: 1014 LVFPFTYSQQQENQEIQLQPPAKRLNCGTNYEIPKIPFLDSGQEMFLRRQQQHQQQQHQQ 1193
Query: 128 QLPLFPHHYLQHQQQQQQSSVVPFAKQQKVSSSTTG---DDASIQLQQSIFDQLFKTAEL 184
QL L PHH Q P A +QK+ + +G D ++ Q QQ+I DQLFK AEL
Sbjct: 1194 QLQLLPHHLQQR----------PMAPKQKMGNPGSGGGEDVSAHQFQQAIIDQLFKAAEL 1343
Query: 185 IEAGNPVQAQGILARLNHQLSPIGNPFQRASFYMKEALQLMLHSNGNNLT--AFSPISFI 242
I+AGNP AQGILARLNHQ+SP+G PFQRA+FY KEALQL+L SN NN +FSP S +
Sbjct: 1344 IDAGNPEHAQGILARLNHQISPMGKPFQRAAFYFKEALQLLLQSNVNNSNNNSFSPTSLL 1523
Query: 243 FKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQEIVLR 302
KIGAYKSFSEISPVLQFANFT NQ+L+EA+E FDRIH+IDFDIGFG QWSSFMQE+ LR
Sbjct: 1524 LKIGAYKSFSEISPVLQFANFTSNQALLEAVEGFDRIHIIDFDIGFGGQWSSFMQELALR 1703
Query: 303 SNGKPSLKITAVVSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLNIESLNLPSCPLPG 362
+ G P+LKITA VSPS +EIEL+FT ENL QYA ++N+ FE +L +ESLN S P P
Sbjct: 1704 NGGAPALKITAFVSPSHHDEIELSFTNENLMQYAGEINMSFELEILTLESLNSVSWPQP- 1880
Query: 363 HFFDSNEAIGVNFPVSSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDRMDVPLPTNVVH 422
EA+ VN P+ SF + PS P+ L F+KQL PKIVVTLD++CDR D P P ++V
Sbjct: 1881 --LRDCEAVVVNLPICSFSNYPSYLPLVLRFVKQLMPKIVVTLDRSCDRTDAPFPQHMVF 2054
Query: 423 VLQCYSALLESLDAVNVNL--DVLQKIERHYIQPTINKIVLSHHNQRDKLPPWRNMFLQS 480
LQ YS LLESLDAV+VN+ DVLQ IE++Y+QP I K+VL +D+ PW+N+ L S
Sbjct: 2055 ALQSYSGLLESLDAVSVNVHPDVLQMIEKYYLQPAIEKLVLGRLRSQDRTLPWKNLLLSS 2234
Query: 481 GFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRKELISVSTWRC 533
GFSP +FSNFTE+QAECLVQR P RGF VE+K +SLVLCWQRK+LISVSTWRC
Sbjct: 2235 GFSPLTFSNFTESQAECLVQRIPGRGFHVEKKQNSLVLCWQRKDLISVSTWRC 2393
Score = 57.4 bits (137), Expect = 1e-08
Identities = 59/196 (30%), Positives = 86/196 (43%), Gaps = 42/196 (21%)
Frame = +3
Query: 18 EKCGGGGMRMEDWEG---------QDQSLLRLIMGDVEDPSAGLNKILQ---NSGYGSQN 65
++CG MEDWE QD S+L+LIM D+ED S GL+K+LQ ++ + Q+
Sbjct: 531 KRCG-----MEDWESVLSESPSQDQDHSILKLIMEDIEDASMGLSKLLQVGTSNSHSQQD 695
Query: 66 VDFHGGFGVLDHQQQQGLTMMDASVQQGNYN-----------------VFPFIPENYNVL 108
V+F+ GF +D Q +++D GN N FPF N +
Sbjct: 696 VEFN-GFSFMDQQS----SVLDPISSNGNNNNFVTSIDSSSVVATTATDFPFGSRGMNQI 860
Query: 109 --PLLDSGQEVFA-----RRHQQQE--TQLPLFPHHYLQHQQQQQ----QSSVVPFAKQQ 155
P S Q + +HQQQ + PH ++ +Q Q Q S V PF Q
Sbjct: 861 QNPNFSSSQVSISHGGLFHQHQQQSIGSLDQKLPHQFVMNQNQAQFMPNPSLVFPFTYSQ 1040
Query: 156 KVSSSTTGDDASIQLQ 171
+ ++ IQLQ
Sbjct: 1041Q------QENQEIQLQ 1070
>BM779515 weakly similar to GP|18087577|gb| AT4g00150/F6N15_20 {Arabidopsis
thaliana}, partial (4%)
Length = 342
Score = 234 bits (597), Expect = 6e-62
Identities = 111/111 (100%), Positives = 111/111 (100%)
Frame = +3
Query: 12 NDDNSEEKCGGGGMRMEDWEGQDQSLLRLIMGDVEDPSAGLNKILQNSGYGSQNVDFHGG 71
NDDNSEEKCGGGGMRMEDWEGQDQSLLRLIMGDVEDPSAGLNKILQNSGYGSQNVDFHGG
Sbjct: 9 NDDNSEEKCGGGGMRMEDWEGQDQSLLRLIMGDVEDPSAGLNKILQNSGYGSQNVDFHGG 188
Query: 72 FGVLDHQQQQGLTMMDASVQQGNYNVFPFIPENYNVLPLLDSGQEVFARRH 122
FGVLDHQQQQGLTMMDASVQQGNYNVFPFIPENYNVLPLLDSGQEVFARRH
Sbjct: 189 FGVLDHQQQQGLTMMDASVQQGNYNVFPFIPENYNVLPLLDSGQEVFARRH 341
>TC88326 similar to GP|18087577|gb|AAL58919.1 AT4g00150/F6N15_20
{Arabidopsis thaliana}, partial (6%)
Length = 744
Score = 152 bits (383), Expect = 4e-37
Identities = 75/86 (87%), Positives = 75/86 (87%)
Frame = +3
Query: 1 MVISENLAHLSNDDNSEEKCGGGGMRMEDWEGQDQSLLRLIMGDVEDPSAGLNKILQNSG 60
MVISENLAHLSNDDNSEEKCGGGGMRMEDWEGQDQSLLRLIMGDVEDPSAGLNKILQNSG
Sbjct: 486 MVISENLAHLSNDDNSEEKCGGGGMRMEDWEGQDQSLLRLIMGDVEDPSAGLNKILQNSG 665
Query: 61 YGSQNVDFHGGFGVLDHQQQQGLTMM 86
YGSQNVDF GF Q LTMM
Sbjct: 666 YGSQNVDFSWGFWCFGSSXQXXLTMM 743
>TC77063 similar to GP|20334379|gb|AAM19210.1 GAI-like protein 1 {Vitis
vinifera}, partial (71%)
Length = 1762
Score = 130 bits (328), Expect = 1e-30
Identities = 98/384 (25%), Positives = 169/384 (43%), Gaps = 10/384 (2%)
Frame = +3
Query: 159 SSTTGDDASIQLQQS---IFDQLFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRAS 215
S+TT ++ Q+ + L AE +E N A+ ++ ++ + ++ +
Sbjct: 546 STTTRPIMVVETQEKGIRLVHSLMACAEAVEQNNLKMAEALVKQIGYLAVSQEGAMRKVA 725
Query: 216 FYMKEALQLMLHSNGNNLTAFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALER 275
Y E L ++ F S + + F E P L+FA+FT NQ+++EA +
Sbjct: 726 TYFAEGLARRIYG------VFPQHSVSDSLQIH--FYETCPNLKFAHFTANQAILEAFQG 881
Query: 276 FDRIHVIDFDIGFGVQWSSFMQEIVLRSNGKPSLKITAVVSPSSCNEIELNFTQENLSQY 335
+HVIDF I G+QW + MQ + LR G P+ ++T + P+S N L L+Q+
Sbjct: 882 KSSVHVIDFSINQGMQWPALMQALALRPGGPPAFRLTGIGPPASDNSDHLQQVGWRLAQF 1061
Query: 336 AKDLNILFEFNVLNIESLNLPSCPLPGHFFDSNEAIGVN--FPVSSFISNPSCFPVALHF 393
A+ +++ FE+ SL + E++ VN F + + P
Sbjct: 1062AQTIHVQFEYRGFVANSLADLDASMLELRSPETESVAVNSVFELHKLNARPGALEKVFSV 1241
Query: 394 LKQLRPKIVVTLDKNCDRMDVPLPTNVVHVLQCYSALLESLDAVNVNLDVLQKIERHYIQ 453
++Q+RP+IV +++ + L YS L +SL+ +V + + Y+
Sbjct: 1242IRQIRPEIVTVVEQEANHNGPAFLDRFTESLHYYSTLFDSLEGSSVEPQD-KAMSEVYLG 1418
Query: 454 PTINKIVLSHHNQR----DKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQR-APVRGFQ 508
I +V R + L WRN F +GFSP + QA L+ A G++
Sbjct: 1419KQICNVVACEGTDRVERHETLNQWRNRFNSAGFSPVHLGSNAFKQASMLLALFAGGDGYK 1598
Query: 509 VERKPSSLVLCWQRKELISVSTWR 532
VE L+L W + LI+ S W+
Sbjct: 1599VEENDGCLMLGWHTRPLIATSAWK 1670
>TC76809 similar to GP|20334379|gb|AAM19210.1 GAI-like protein 1 {Vitis
vinifera}, partial (72%)
Length = 2764
Score = 123 bits (308), Expect = 2e-28
Identities = 99/379 (26%), Positives = 169/379 (44%), Gaps = 24/379 (6%)
Frame = +2
Query: 178 LFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRASFYMKEALQLMLHSNGNNLTAFS 237
L AE I+ N A+ ++ ++ S ++ + Y +AL ++ N T S
Sbjct: 1376 LMACAEAIQQKNLKLAEALVKHISLLASLQTGAMRKVASYFAQALARRIYGNPEE-TIDS 1552
Query: 238 PISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQ 297
S I + Y+S SP L+FA+FT NQ+++EA R+HVIDF + G+QW + MQ
Sbjct: 1553 SFSEILHMHFYES----SPYLKFAHFTANQAILEAFAGAGRVHVIDFGLKQGMQWPALMQ 1720
Query: 298 EIVLRSNGKPSLKITAVVSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLNIESLNLPS 357
+ LR G P+ ++T + P + N L L+Q A+ + + FEF S+
Sbjct: 1721 ALALRPGGPPTFRLTGIGPPQADNTDALQQVGWKLAQLAQTIGVQFEFRGFVCNSI---- 1888
Query: 358 CPLPGHFFD--SNEAIGVN--FPVSSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDRMD 413
L + + EA+ VN F + + ++ P L+ +K++ PKIV +++ +
Sbjct: 1889 ADLDPNMLEIRPGEAVAVNSVFELHTMLARPGSVEKVLNTVKKINPKIVTIVEQEANHNG 2068
Query: 414 VPLPTNVVHVLQCYSALLESLDAVN---------------VNLDVLQKIERHYIQPTINK 458
L YS+L +SL+ N + D+L + Y+ I
Sbjct: 2069 PVFVDRFTEALHYYSSLFDSLEGSNSSSNNSNSNSTGLGSPSQDLL--MSEIYLGKQICN 2242
Query: 459 IV----LSHHNQRDKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQR-APVRGFQVERKP 513
+V + + + L WR+ +GF P + QA L+ A G++VE
Sbjct: 2243 VVAYEGVDRVERHETLTQWRSRMGSAGFEPVHLGSNAFKQASTLLALFAGGDGYRVEENN 2422
Query: 514 SSLVLCWQRKELISVSTWR 532
L+L W + LI+ S W+
Sbjct: 2423 GCLMLGWHTRSLIATSAWK 2479
>TC79190 similar to PIR|E96542|E96542 scarecrow-like protein [imported] -
Arabidopsis thaliana, partial (64%)
Length = 1669
Score = 122 bits (305), Expect = 4e-28
Identities = 95/382 (24%), Positives = 177/382 (45%), Gaps = 25/382 (6%)
Frame = +2
Query: 176 DQLFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRASFYMKEALQLMLHSNGNNL-- 233
+ L+ A+ ++ + + ++ L +S G+P +R YM EAL + S+G+ +
Sbjct: 422 EMLYTCAKAVDENDIETIEWMVTELRKIVSVSGSPIERLGAYMLEALVSKIASSGSTIYK 601
Query: 234 -------TAFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDI 286
T +S++ + EI P +F + N ++ EA++ + +H+IDF I
Sbjct: 602 SLKCSEPTGNELLSYMHVL------YEICPYFKFGYMSANGAIAEAMKEENEVHIIDFQI 763
Query: 287 GFGVQWSSFMQEIVLRSNGKPSLKITAVVSPSSCNEI--ELNFTQENLSQYAKDLNILFE 344
G G QW S +Q + R G P ++IT + S N ++ E L A+ ++ FE
Sbjct: 764 GQGTQWVSLIQALARRPGGPPKIRITGIDDSYSSNVRGGGVDIVGEKLLTLAQSCHVPFE 943
Query: 345 FNVLNIESLNLPS-CPLPGHFFDSNEAIGVNFPV------SSFISNPSCFPVALHFLKQL 397
F+ + + PS L NEA+ VNF + ++ + L K +
Sbjct: 944 FHAVRV----YPSEVRLEDFELRPNEAVAVNFAIMLHHVPDESVNIHNHRDRLLRLAKHM 1111
Query: 398 RPKIVVTLDKNCDRMDVPLPTNVVHVLQCYSALLESLDAVNVNLDVLQKI--ERHYIQPT 455
PK+V +++ + + P + + YSA+ ES+D V + D ++I E+H +
Sbjct: 1112SPKVVTLVEQEFNTNNAPFLQRFLETMNYYSAVYESIDVV-LPRDHKERINVEQHCLARE 1288
Query: 456 INKIVLSHHNQR----DKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRG-FQVE 510
+ +V +R + L WR F +GF+P+ S+F + + L++ RG + +E
Sbjct: 1289VVNLVACEGEERVERHELLSKWRMRFTMAGFTPYPLSSFINSSIKNLLE--SYRGHYTLE 1462
Query: 511 RKPSSLVLCWQRKELISVSTWR 532
+ +L L W ++LI+ WR
Sbjct: 1463ERDGALFLGWMNQDLIASCAWR 1528
>TC77707 weakly similar to GP|8777405|dbj|BAA96995.1 SCARECROW gene
regulator-like {Arabidopsis thaliana}, partial (65%)
Length = 2161
Score = 110 bits (274), Expect = 2e-24
Identities = 102/424 (24%), Positives = 177/424 (41%), Gaps = 28/424 (6%)
Frame = +2
Query: 136 YLQHQ------QQQQQSSVVPFAKQQKVSSSTTGDDASIQLQQS--IFDQLFKTAELIEA 187
Y+Q Q +Q + V K++K+ +D+S+Q S + L A+ +
Sbjct: 743 YIQSQPSHVTSSSRQSNEAVHVEKRRKLE-----EDSSLQGFPSGDLKQLLIACAKAMAE 907
Query: 188 GNPVQAQGILARLNHQLSPIGNPFQRASFYMKEALQLMLHSNGNNLTAFSPISFIFKIGA 247
N ++ + +S G P QR YM E L ++GN++ + + G
Sbjct: 908 NNTELFDRLIETARNAVSINGEPIQRLGAYMVEGLVARTEASGNSI--YHALKCREPEGE 1081
Query: 248 -----YKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQEIVLR 302
+ EI P L+F N ++ EA D IH+IDF I G QW + +Q + R
Sbjct: 1082 ELLTYMQLLFEICPYLKFGYMAANGAIAEACRNEDHIHIIDFQIAQGTQWMTLLQALAAR 1261
Query: 303 SNGKPSLKITAVVSPSS--CNEIELNFTQENLSQYAKDLNILFEFNVLNIESLNLPSCPL 360
G P ++IT + P S L E LS +K I EF+ + + ++ L
Sbjct: 1262 PGGAPHVRITGIDDPVSKYARGKGLEVVGERLSLMSKKFGIPVEFHGIPVFGPDVTRDML 1441
Query: 361 PGHFFDSNEAIGVNFPV--------SSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDRM 412
EA+ VNFP+ S ++NP L +K L PK+V +++ +
Sbjct: 1442 D---IRHGEALAVNFPLQLHHTADESVDVNNPR--DGLLRLVKSLSPKVVTLVEQESNTN 1606
Query: 413 DVPLPTNVVHVLQCYSALLESLD-AVNVNLDVLQKIERHYIQPTINKIVLSHHNQR---- 467
P + L Y A+ ES+D ++ N +E+H + I ++ +R
Sbjct: 1607 TTPFFNRFIETLDYYLAIFESIDVTLSRNSKERINVEQHCLARDIVNVIACEGKERVERH 1786
Query: 468 DKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRKELIS 527
+ W++ +GF S++ + L+ R + + K +++L W+ + LIS
Sbjct: 1787 ELFGKWKSRLTMAGFRQCPLSSYVNSVIRSLL-RCYSEHYTLVEKDGAMLLGWKSRNLIS 1963
Query: 528 VSTW 531
S W
Sbjct: 1964 ASAW 1975
>TC77896 weakly similar to GP|22087131|gb|AAM90848.1 hairy meristem {Petunia
x hybrida}, partial (8%)
Length = 258
Score = 110 bits (274), Expect = 2e-24
Identities = 57/83 (68%), Positives = 65/83 (77%), Gaps = 2/83 (2%)
Frame = +3
Query: 211 FQRASFYMKEALQLMLHSNGNNLT--AFSPISFIFKIGAYKSFSEISPVLQFANFTCNQS 268
F RA Y KEALQL+L SN NN +FS + KIGAYKSFS+ISPVLQFANFT NQ+
Sbjct: 9 FSRAGCYCKEALQLLLQSNVNNSNNNSFSGTRLLLKIGAYKSFSQISPVLQFANFTSNQA 188
Query: 269 LIEALERFDRIHVIDFDIGFGVQ 291
L+EA+ERFDRIH+ID DIGFG Q
Sbjct: 189 LLEAVERFDRIHIIDCDIGFGGQ 257
>TC86258 similar to GP|16930433|gb|AAL31902.1 AT4g17230/dl4650c {Arabidopsis
thaliana}, partial (62%)
Length = 2237
Score = 109 bits (272), Expect = 3e-24
Identities = 92/376 (24%), Positives = 163/376 (42%), Gaps = 20/376 (5%)
Frame = +3
Query: 178 LFKTAELIEAGNPVQAQGILAR-LNHQLSPIGNPFQRASFYMKEALQLMLHSNGN----N 232
L A+ + G+ A+G + L +S G+P QR S Y+ E L+ L +G+ +
Sbjct: 873 LLLCAQAVSDGDIPTARGWMDNVLVKMVSVAGDPIQRLSAYLLEGLRARLELSGSLIYKS 1052
Query: 233 LTAFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQW 292
L P S + + +I P +FA + N + EA+ RIH+IDF I G QW
Sbjct: 1053 LKCEQPTSKELMTYMHMLY-QICPYFKFAYISANAVISEAMANESRIHIIDFQIAQGTQW 1229
Query: 293 SSFMQEIVLRSNGKPSLKITAVVSPSSCNEI--ELNFTQENLSQYAKDLNILFEFNVLNI 350
++ + R G P ++IT V S + L E LS +A+ +LFEF+
Sbjct: 1230 QMLIEALARRPGGPPFIRITGVDDSQSFHARGGGLQIVGEQLSNFARSRGVLFEFH---- 1397
Query: 351 ESLNLPSCPLPGHFF--DSNEAIGVNFPVS------SFISNPSCFPVALHFLKQLRPKIV 402
S + C + EA+ VNFP S +S + L +K L PK+V
Sbjct: 1398 -SAAMSGCEVQRENLRVSPGEALAVNFPFSLHHMPDESVSIENHRDRLLRLVKSLSPKVV 1574
Query: 403 VTLDKNCDRMDVPLPTNVVHVLQCYSALLESLDAVNVNLDVLQ-KIERHYIQPTINKIV- 460
+++ + P V + Y+A+ ES+D D + +E++ + I ++
Sbjct: 1575 TLVEQESNTNTSPFFQRFVETMDFYTAMFESIDVACTKDDKKRISVEQNCVARDIVNMIA 1754
Query: 461 ---LSHHNQRDKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLV 517
+ + + WR+ F +GF S+ + +++ + + +E + +L
Sbjct: 1755 CEGIERVERHEVFGKWRSRFSMAGFRQCQLSSSVMHSVQNMLKDFH-QNYWLEHRDGALY 1931
Query: 518 LCWQRKELISVSTWRC 533
L W ++ + + S W C
Sbjct: 1932 LGWMKRAMATSSAWMC 1979
>TC77716 similar to GP|20465587|gb|AAM20276.1 unknown protein {Arabidopsis
thaliana}, partial (63%)
Length = 2392
Score = 106 bits (264), Expect = 3e-23
Identities = 102/398 (25%), Positives = 173/398 (42%), Gaps = 17/398 (4%)
Frame = +3
Query: 152 AKQQKVSSSTTGDDASIQLQQSIFDQLFKTAELIEAGNPVQAQGILARLNHQLSPIGNPF 211
+ +SS+++ + S ++ L++ A + GN V+A ++ L +S G P
Sbjct: 825 SSDSNLSSTSSTKEISQNFPRTPKQLLYECASALSEGNEVKASAMIDDLRQLVSIQGEPS 1004
Query: 212 QRASFYMKEALQLMLHSNGN----NLTAFSPISFIFKIGAYKSFSEISPVLQFANFTCNQ 267
R + YM E L L S+G L P S ++ A + E+ P +F N
Sbjct: 1005 DRIAAYMVEGLAARLASSGKCIYKALKCKEPPSSD-RLAAMQILFEVCPCFKFGFIAANG 1181
Query: 268 SLIEALERFDRIHVIDFDIGFGVQWSSFMQEIVLRSNGKPSLKITAVVSPSSCNEI--EL 325
++ EA++ ++H+IDFDI G Q+ + +Q + R P +++T V P S L
Sbjct: 1182 AISEAIKDDKKVHIIDFDINQGSQYITLIQTLASRPGKPPYVRLTGVDDPESVQRSVGGL 1361
Query: 326 NFTQENLSQYAKDLNILFEFN-VLNIESLNLPSC--PLPGHFFDSNEAIGVNFPVSSFIS 382
+ + L + A+ L + FEF V + S+ PS PG N A ++ +S
Sbjct: 1362 SIIGQRLEKLAEVLGLPFEFRAVASRSSIVTPSMLNCRPGEALVVNFAFQLHHMPDESVS 1541
Query: 383 NPSCFPVALHFLKQLRPKIVVTLDKNCDRMDVPLPTNVVHVLQCYSALLESLDAV--NVN 440
+ L +K L PK+V ++++ + P V YSA+ ESLDA +
Sbjct: 1542 TVNERDQLLRMVKSLNPKLVTVVEQDVNTNTSPFLPRFVEAYNYYSAVFESLDATLPRES 1721
Query: 441 LDVLQKIERHYIQPTINKIVLSHHNQR----DKLPPWRNMFLQSGF--SPFSFSNFTEAQ 494
D + +ER + I ++ R + WR +GF SP S +N EA
Sbjct: 1722 QDRV-NVERQCLARDIVNVIACEGEDRIERYEVAGKWRARMKMAGFTSSPMS-TNVKEAI 1895
Query: 495 AECLVQRAPVRGFQVERKPSSLVLCWQRKELISVSTWR 532
E + Q +++ + L W+ K LI S W+
Sbjct: 1896 RELIRQYCD--KYKIIEEMGGLHFGWEDKNLIVASAWK 2003
>BM813961 homologue to GP|13365610|db SCARECROW {Pisum sativum}, partial
(21%)
Length = 550
Score = 89.0 bits (219), Expect = 4e-18
Identities = 54/171 (31%), Positives = 88/171 (50%), Gaps = 3/171 (1%)
Frame = -2
Query: 178 LFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRASFYMKEALQLMLHSNGNNLTAFS 237
L + AE + A N QA +L ++ +P G QR + Y EA+ L S+ + A
Sbjct: 543 LLQCAEAVSAENLDQANKMLLEISQLSTPFGTSAQRVAAYFSEAISARLVSSCLRIYATL 364
Query: 238 PISFIFK---IGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSS 294
P + A++ F+ ISP ++F++FT NQ++ EA +R +R+H+ID DI G+QW
Sbjct: 363 PPHTLHNQKVASAFQVFNGISPFVKFSHFTANQAIQEAFDREERVHIIDLDIMQGLQWPG 184
Query: 295 FMQEIVLRSNGKPSLKITAVVSPSSCNEIELNFTQENLSQYAKDLNILFEF 345
+ R G P +++T + + L T + LS +A L + FEF
Sbjct: 183 LFHILASRPGGPPYVRLTGLGTSME----TLEATGKRLSDFASKLGLPFEF 43
>TC90344 weakly similar to PIR|H86282|H86282 protein F10B6.34 [imported] -
Arabidopsis thaliana, partial (28%)
Length = 1170
Score = 87.4 bits (215), Expect = 1e-17
Identities = 83/322 (25%), Positives = 139/322 (42%), Gaps = 17/322 (5%)
Frame = +1
Query: 41 IMGDVEDPSA--GLNKILQNSGYGSQNVDFHGGFGVLDHQQQQGLTMMDASVQQGNYNVF 98
I+ D PS ++ +L N GY ++ + H Q Q L +++++ + ++
Sbjct: 175 IVSDSSSPSKTKDIDGLLANVGYKVRSSELH--------QVAQNLERLESAIVNSSSDIS 330
Query: 99 PFIPENYNVLPLLDSGQEVFARRHQQQET--------QLPLFPHHYLQHQQQQQQSSVVP 150
F + + P D G V + T QLP QH Q S+V
Sbjct: 331 QFASDTVHYDPS-DIGNWVDNLLSEFDHTASLPYDFSQLPDLSAPPPQHHHHQSSSTV-- 501
Query: 151 FAKQQKVSSSTTGDDASIQLQQSIFDQLFKTAELIEAGNPVQA-------QGILARLNHQ 203
+ +D++I+L L A+ ++ GN A QG+LA +N
Sbjct: 502 ---------NNVEEDSAIKLVH----MLMTCADSVQRGNLSLAGSLIEGMQGLLANMNTN 642
Query: 204 LSPIGNPFQRASFYMKEALQLMLHSNGNNLTAFSPISFIFKIGAYKSFSEISPVLQFANF 263
S IG + + Y +AL + N +S Y + E P L+FA+F
Sbjct: 643 -SGIG----KVAGYFIDALNRRIFGQNN---VSHQVSLYENDVLYHHYYEACPYLKFAHF 798
Query: 264 TCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQEIVLRSNGKPSLKITAVVSPSSCNEI 323
T NQ+++EA D +HVIDF++ G+QW + +Q + LR G P L++T + PS +
Sbjct: 799 TANQAILEAFNGHDCVHVIDFNLMHGLQWPALIQALALRPGGPPFLRLTGIGPPSPDDRD 978
Query: 324 ELNFTQENLSQYAKDLNILFEF 345
L L++ A+ +N+ F F
Sbjct: 979 NLREIGLRLAELARSVNVRFAF 1044
>TC82600 similar to GP|9757937|dbj|BAB08425.1 SCARECROW gene regulator-like
protein {Arabidopsis thaliana}, partial (51%)
Length = 1141
Score = 84.3 bits (207), Expect = 1e-16
Identities = 58/203 (28%), Positives = 102/203 (49%), Gaps = 10/203 (4%)
Frame = +2
Query: 154 QQKVSSSTTGDDASIQLQQSIFDQLFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQR 213
+ + + T G++++ + L + AE + N A +L + SP G +R
Sbjct: 446 EHETETETEGEEST---GLKLLGLLLQCAECVAMDNLDFANDLLPEITELSSPFGTSPER 616
Query: 214 ASFYMKEALQLMLHSNGNNLTAFSPISF----------IFKIGAYKSFSEISPVLQFANF 263
Y +ALQ + S+ L ++SP++ IF A++S++ +SP+++F++F
Sbjct: 617 VGAYFAQALQARVVSSC--LGSYSPLTAKSVTLNQSQRIFN--AFQSYNSVSPLVKFSHF 784
Query: 264 TCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQEIVLRSNGKPSLKITAVVSPSSCNEI 323
T NQ++ +AL+ DR+H+ID DI G+QW + RS S++IT S S
Sbjct: 785 TANQAIFQALDGEDRVHIIDLDIMQGLQWPGLFHILASRSKKIRSVRITGFGSSSEL--- 955
Query: 324 ELNFTQENLSQYAKDLNILFEFN 346
L T L+ +A L + FEF+
Sbjct: 956 -LESTGRRLADFASSLGLPFEFH 1021
>TC78035 similar to GP|8778540|gb|AAF79548.1| F22G5.9 {Arabidopsis thaliana},
partial (24%)
Length = 2694
Score = 82.8 bits (203), Expect = 3e-16
Identities = 77/320 (24%), Positives = 133/320 (41%), Gaps = 19/320 (5%)
Frame = +3
Query: 182 AELIEAGNPVQAQGILARLNHQLSPIGNPFQRASFYMKEALQLMLHSNGNNLTAFSPISF 241
A+ + A + A +L ++ + SP G+ QR + Y A++ + G
Sbjct: 1212 AQAVSANDNRTANELLKQIRNHSSPSGDASQRMAHYFANAIEARMVGAGTGTQILYMSQK 1391
Query: 242 IFK----IGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQ 297
+F + AY+ F P +FA+F N+ +++ E+ + +H+IDF I +G QW ++
Sbjct: 1392 MFSAADFLKAYQVFISACPFKKFAHFFANKMILKTAEKAETLHIIDFGILYGFQWPILIK 1571
Query: 298 EIVLRSNGKPSLKITAVVSPSS----CNEIELNFTQENLSQYAKDLNILFEFNVLNIESL 353
+ G P L+IT + P + IE T L+ Y + N+ FE+ I S
Sbjct: 1572 FLSKVEGGPPKLRITGIEYPQAGFRPAERIE--ETGRRLANYCERFNVSFEYKA--IPSR 1739
Query: 354 NLPSCPLPGHFFDSNEAIGVNFPV------SSFISNPSCFPVALHFLKQLRPKIVVTLDK 407
N + + SNE + VN V I S L ++++ P I V
Sbjct: 1740 NWETIQIEDLNIKSNEVVAVNCLVRFKNLHDETIDVNSPKDAVLKLIRKINPHIFVQSIV 1919
Query: 408 NCDRMDVPLPTNVVHVLQCYSALLESLDA-VNVNLDVLQKIERHYIQPTINKIVLSHHNQ 466
N T L YSA+ + D ++ + IER ++ I +V +
Sbjct: 1920 NGSYNAPFFSTRFKESLFHYSAMFDMYDTLISRENEWRSMIEREFLGREIMNVVACEGFE 2099
Query: 467 RDKLP----PWRNMFLQSGF 482
R + P W+ L++GF
Sbjct: 2100 RVERPETYKQWQVRNLRAGF 2159
>BI308453 weakly similar to PIR|T02736|T0 probable SCARECROW gene regulator
[imported] - Arabidopsis thaliana, partial (11%)
Length = 712
Score = 81.3 bits (199), Expect = 9e-16
Identities = 48/172 (27%), Positives = 83/172 (47%), Gaps = 3/172 (1%)
Frame = +3
Query: 178 LFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRASFYMKEALQLMLHSNGNNL---T 234
L A+ I + A +L ++ SP G+ QR + + AL+ L G+++
Sbjct: 195 LMLCAQSISCNDISNANQLLNQIKKHSSPTGDGTQRLAHFFGNALEARLAGTGSHVYRAL 374
Query: 235 AFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSS 294
+ S + AY+ +S P + A N +++ + + +H+IDF +G+G +W
Sbjct: 375 SSKKKSAADMVKAYQVYSSACPFEKLAIMFSNDAILNVAKETESLHIIDFGVGYGFKWLG 554
Query: 295 FMQEIVLRSNGKPSLKITAVVSPSSCNEIELNFTQENLSQYAKDLNILFEFN 346
F+ + RS G P L+IT + P+S + N T LS Y K N+ FE+N
Sbjct: 555 FIYRLSKRSGGPPKLRITGIDLPNSLERV--NETGLRLSSYCKRFNVPFEYN 704
>CA919642 similar to PIR|E85433|E854 SCARECROW-like protein [imported] -
Arabidopsis thaliana, partial (12%)
Length = 661
Score = 78.2 bits (191), Expect = 7e-15
Identities = 41/117 (35%), Positives = 64/117 (54%), Gaps = 3/117 (2%)
Frame = -2
Query: 420 VVHVLQCYSALLESLDA---VNVNLDVLQKIERHYIQPTINKIVLSHHNQRDKLPPWRNM 476
VV+ L+ YS +LESLDA + ++IE ++P KI+ + + PWR
Sbjct: 489 VVNSLEFYSMMLESLDASVAAGGGGEWARRIEMLLLRP---KIIAAVEAAGRRTTPWREA 319
Query: 477 FLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRKELISVSTWRC 533
F +G P S F + QAECL+ + +RGF V ++ + LVL W + +++ S WRC
Sbjct: 318 FYGAGMRPVQLSQFADFQAECLLAKVQIRGFHVAKRQAELVLFWHERAMVATSAWRC 148
>TC89212 weakly similar to PIR|T02531|T02531 probable SCARECROW gene
regulator At2g37650 [imported] - Arabidopsis thaliana,
partial (37%)
Length = 1266
Score = 70.9 bits (172), Expect = 1e-12
Identities = 75/339 (22%), Positives = 139/339 (40%), Gaps = 15/339 (4%)
Frame = +1
Query: 208 GNPFQRASFYMKEALQLMLHSNGNNLTAFSPISFIFKIGAYKSFSEISPVLQFANFTCNQ 267
G+ QR + Y L++ L + + + + AYK F SP+ + N +
Sbjct: 4 GDGLQRLAHYFANGLEIRLAAETPSYQPLDVATAGDMLKAYKLFVTASPLQRVTNTLLTK 183
Query: 268 SLIEALERFDRIHVIDFDIGFGVQWSSFMQEIVLRSNGKPSLKITAV--VSPSSCNEIEL 325
++ + ++ +HVIDF I +G QW ++ + LR G P L+IT + P +
Sbjct: 184 TIFKIVKNESSVHVIDFGICYGFQWPCLVRRLSLRPGGPPKLRITGIELPQPGFRPTERV 363
Query: 326 NFTQENLSQYAKDLNILFEFNVLNIESLNLPSCPLPGHFFDSNEAIGV-------NFPVS 378
T L++Y K N+ FE+N + + + L D NE V N P
Sbjct: 364 EETGRRLAKYCKKFNVPFEYNFI---AQKWETVCLEDLKIDRNEITLVSCLYRLKNLPDE 534
Query: 379 SFISNPSCFPVALHFLKQLRPKIVVTLDKNCDRMDVPLPTNVVHVLQCYSALLESLDAVN 438
+ N V L ++++ PKI N T L +S+L + +A N
Sbjct: 535 TVALNCPREAV-LKLIRKINPKIFFHGVVNGSYSAPFFLTRFKEALYHFSSLFDMFEA-N 708
Query: 439 VNLDVLQK--IERHYIQPTINKIVLSHHNQRDKLP----PWRNMFLQSGFSPFSFSNFTE 492
V + Q+ +ER ++ +R + P W+ ++GF F +
Sbjct: 709 VPCEDPQRLMLERGLFGRDAINVIACEGAERVERPETYKQWQVRNKRAGFRQIRFDSDLV 888
Query: 493 AQAECLVQRAPVRGFQVERKPSSLVLCWQRKELISVSTW 531
+ + +V++ + F V+ ++ W+ + L ++S W
Sbjct: 889 NETKAMVKKEYHKDFVVDVDGKWVLQGWKGRILNALSAW 1005
>TC80602 similar to GP|14517552|gb|AAK62666.1 F17J6.12/F17J6.12 {Arabidopsis
thaliana}, partial (31%)
Length = 1190
Score = 70.5 bits (171), Expect = 2e-12
Identities = 43/142 (30%), Positives = 68/142 (47%), Gaps = 4/142 (2%)
Frame = +2
Query: 182 AELIEAGNPVQAQGILARLNHQLSPIGNPFQRASFYMKEALQLMLHSNGNN----LTAFS 237
A+ I + + AQ ++ L +S G P QR YM E L L ++G++ L
Sbjct: 749 AKAIADNDLLMAQWLMDELRQMVSVSGEPIQRLGAYMLEGLVARLSASGSSIYKSLRCKE 928
Query: 238 PISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQ 297
P S + E+ P +F + N ++ EA++ R+H+IDF I G QW S +Q
Sbjct: 929 PESAEL-LSYMNILYEVCPYFKFGYMSANGAIAEAMKNEARVHIIDFQIAQGSQWISLIQ 1105
Query: 298 EIVLRSNGKPSLKITAVVSPSS 319
R G P ++IT + P+S
Sbjct: 1106AFAARPGGPPHIRITGIDDPTS 1171
>TC83571 similar to PIR|T10552|T10552 hypothetical protein T12G13.90 -
Arabidopsis thaliana, partial (19%)
Length = 767
Score = 68.2 bits (165), Expect = 8e-12
Identities = 32/80 (40%), Positives = 54/80 (67%)
Frame = +1
Query: 245 IGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQEIVLRSN 304
+ A++ ++SP ++F +FT NQ++IEA+ R+HVID+DI GVQW+S +Q + +N
Sbjct: 502 LAAFQLLQDMSPYVKFGHFTANQAIIEAVAHERRVHVIDYDIMEGVQWASLIQSLASNNN 681
Query: 305 GKPSLKITAVVSPSSCNEIE 324
G P L+ITA+ + E++
Sbjct: 682 G-PHLRITALSRTGTGPEVK 738
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.136 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,116,734
Number of Sequences: 36976
Number of extensions: 252643
Number of successful extensions: 1880
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 1702
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1816
length of query: 533
length of database: 9,014,727
effective HSP length: 101
effective length of query: 432
effective length of database: 5,280,151
effective search space: 2281025232
effective search space used: 2281025232
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC121238.1


