

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121237.9 + phase: 0 /pseudo
(652 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
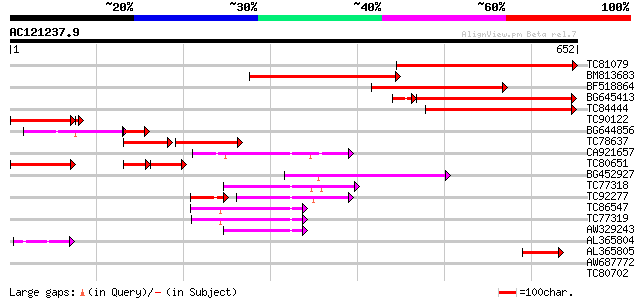
Score E
Sequences producing significant alignments: (bits) Value
TC81079 similar to GP|9758223|dbj|BAB08722.1 FRO2 homolog {Arabi... 387 e-108
BM813683 weakly similar to GP|15341529|gb ferric-chelate reducta... 322 3e-88
BF518864 weakly similar to GP|9758223|dbj| FRO2 homolog {Arabido... 320 8e-88
BG645413 similar to GP|15341529|gb ferric-chelate reductase {Pis... 223 2e-60
TC84444 similar to GP|15341529|gb|AAK95654.1 ferric-chelate redu... 208 5e-54
TC90122 148 1e-35
BG644856 similar to GP|18087544|gb| AT5g23980/MZF18_14 {Arabidop... 104 6e-33
TC78637 similar to GP|8978270|dbj|BAA98161.1 FRO2-like protein; ... 83 5e-26
CA921657 similar to PIR|T03826|T03 cytochrome b245 beta chain ho... 95 1e-19
TC80651 similar to GP|15341529|gb|AAK95654.1 ferric-chelate redu... 73 4e-15
BG452927 weakly similar to GP|20268772|gb putative FRO2; NADPH o... 79 5e-15
TC77318 similar to GP|16549089|dbj|BAB70751. respiratory burst o... 79 7e-15
TC92277 similar to GP|16549089|dbj|BAB70751. respiratory burst o... 72 5e-14
TC86547 similar to GP|20805911|gb|AAM28891.1 NADPH oxidase {Nico... 72 9e-13
TC77319 similar to GP|16549089|dbj|BAB70751. respiratory burst o... 68 1e-11
AW329243 similar to GP|16549089|db respiratory burst oxidase hom... 55 6e-08
AL365804 weakly similar to GP|18377668|gb| putative FRO1 and FRO... 48 1e-05
AL365805 similar to GP|18377668|gb| putative FRO1 and FRO2 prote... 45 1e-04
AW687772 similar to GP|16549089|db respiratory burst oxidase hom... 40 0.002
TC80702 similar to GP|8978271|dbj|BAA98162.1 FRO1-like protein; ... 40 0.003
>TC81079 similar to GP|9758223|dbj|BAB08722.1 FRO2 homolog {Arabidopsis
thaliana}, partial (17%)
Length = 929
Score = 387 bits (993), Expect = e-108
Identities = 180/208 (86%), Positives = 193/208 (92%)
Frame = +3
Query: 445 AYITREKQEPSKDTQKQIQTIWFKTNLSDCPISAVLGPNNWLWLGAIITSSFVMFLLFLG 504
A +TREK+EPS+DTQKQIQTIWFKTNLSD PISAVLGPNNWLWLGAIITSSF+MFLL LG
Sbjct: 6 AXMTREKEEPSRDTQKQIQTIWFKTNLSDSPISAVLGPNNWLWLGAIITSSFIMFLLLLG 185
Query: 505 IVTRYYIYPIENNSGEVYNWTYGVMWYMFSFCSCICICSSVVFLWLKRLNKLENKHIMNV 564
IVTRYYIYPIENN+GEVYNWT VMWYMF C+C+CICSSVVFLW KR N +ENK IMNV
Sbjct: 186 IVTRYYIYPIENNTGEVYNWTSKVMWYMFLLCACVCICSSVVFLWCKRQNTIENKQIMNV 365
Query: 565 EVSTPARSPGSWIYGSERELESLPHQSLVQATNVHFGSRPDLKKILFECEGKDVGVMTCG 624
EV TP RSPGSWIYGSERELESLPHQSL+QATNVHFG+RPDLKKILFEC+ KDVGVM CG
Sbjct: 366 EVPTPTRSPGSWIYGSERELESLPHQSLLQATNVHFGARPDLKKILFECKDKDVGVMVCG 545
Query: 625 PRKMRHEVARICASGLADNLHFESISFN 652
PRK+RHEVA+ICASGLADNLHFESISFN
Sbjct: 546 PRKLRHEVAKICASGLADNLHFESISFN 629
>BM813683 weakly similar to GP|15341529|gb ferric-chelate reductase {Pisum
sativum}, partial (21%)
Length = 523
Score = 322 bits (825), Expect = 3e-88
Identities = 158/174 (90%), Positives = 165/174 (94%)
Frame = +1
Query: 276 LLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHPFTVTSSCNMETNYLSVAIKNV 335
LLPCDALELNFSK+PSLYYNPTSLVFINVPKVSKLQWHPFTV+SSCN+ETN LSV IKNV
Sbjct: 1 LLPCDALELNFSKDPSLYYNPTSLVFINVPKVSKLQWHPFTVSSSCNLETNCLSVTIKNV 180
Query: 336 GSWSNKLYQELSSSSLDHLNISVEGPYGPHTAQFLRHEQIVMVSGGSGVTPFISIIRDLI 395
GSWSNKLYQELSSSSLDHLN+SVEGPYGPH+AQFLRHEQI MVSGGSG+TPFISIIRDLI
Sbjct: 181 GSWSNKLYQELSSSSLDHLNVSVEGPYGPHSAQFLRHEQIAMVSGGSGITPFISIIRDLI 360
Query: 396 FQSQQQEFQPPKLLLVCIFKNYADLTMLDLMLPISGLKTRISQLQLQIEAYITR 449
FQSQQQEFQPPKLLLVCIFKNY DL MLDLMLP SG T+ISQL LQIEAYITR
Sbjct: 361 FQSQQQEFQPPKLLLVCIFKNYVDLAMLDLMLPGSGSTTQISQLPLQIEAYITR 522
>BF518864 weakly similar to GP|9758223|dbj| FRO2 homolog {Arabidopsis
thaliana}, partial (13%)
Length = 469
Score = 320 bits (821), Expect = 8e-88
Identities = 154/156 (98%), Positives = 155/156 (98%)
Frame = +2
Query: 417 YADLTMLDLMLPISGLKTRISQLQLQIEAYITREKQEPSKDTQKQIQTIWFKTNLSDCPI 476
YADLTMLDLMLPISGLKTRISQLQLQIEAYITREKQEPSKDTQKQIQTIWFKTNLSDCPI
Sbjct: 2 YADLTMLDLMLPISGLKTRISQLQLQIEAYITREKQEPSKDTQKQIQTIWFKTNLSDCPI 181
Query: 477 SAVLGPNNWLWLGAIITSSFVMFLLFLGIVTRYYIYPIENNSGEVYNWTYGVMWYMFSFC 536
SAVLGPNNWLWLGAIITSSFVMFLLFLGIVTRYYIYPIENNSGEVYNWTYGVMWYMFSFC
Sbjct: 182 SAVLGPNNWLWLGAIITSSFVMFLLFLGIVTRYYIYPIENNSGEVYNWTYGVMWYMFSFC 361
Query: 537 SCICICSSVVFLWLKRLNKLENKHIMNVEVSTPARS 572
SCICICSSVVFLWLKRLNKLENKHIMNVEVST +RS
Sbjct: 362 SCICICSSVVFLWLKRLNKLENKHIMNVEVSTTSRS 469
>BG645413 similar to GP|15341529|gb ferric-chelate reductase {Pisum sativum},
partial (29%)
Length = 815
Score = 223 bits (569), Expect(2) = 2e-60
Identities = 102/184 (55%), Positives = 140/184 (75%)
Frame = -2
Query: 468 KTNLSDCPISAVLGPNNWLWLGAIITSSFVMFLLFLGIVTRYYIYPIENNSGEVYNWTYG 527
K N +D PI ++LGPN WLWLGAII+SSF++FL+ +GI+TRYYI+PI++N+ +++++
Sbjct: 736 KANPTDAPIHSMLGPNTWLWLGAIISSSFIIFLIIIGIITRYYIFPIDHNTNKIFSYPLR 557
Query: 528 VMWYMFSFCSCICICSSVVFLWLKRLNKLENKHIMNVEVSTPARSPGSWIYGSERELESL 587
V + C + + +SV +W K+ N E K I N+E S+P SP S IY +RELESL
Sbjct: 556 VFLNVLVICVSVVVVASVAVIWNKKQNAKEAKQIQNLEGSSPTVSPSSMIYNVDRELESL 377
Query: 588 PHQSLVQATNVHFGSRPDLKKILFECEGKDVGVMTCGPRKMRHEVARICASGLADNLHFE 647
P+QSLVQATNVH+G+RPDLK++LFE +G VGV+ GP++MR EVA IC+SGL +NLHFE
Sbjct: 376 PYQSLVQATNVHYGTRPDLKRLLFEMKGSSVGVLVSGPKQMRQEVASICSSGLVENLHFE 197
Query: 648 SISF 651
SISF
Sbjct: 196 SISF 185
Score = 27.7 bits (60), Expect(2) = 2e-60
Identities = 14/28 (50%), Positives = 19/28 (67%)
Frame = -3
Query: 441 LQIEAYITREKQEPSKDTQKQIQTIWFK 468
LQIEAYI R+++ S D+ QT+W K
Sbjct: 813 LQIEAYIPRDREFKS-DSSIHPQTLWLK 733
>TC84444 similar to GP|15341529|gb|AAK95654.1 ferric-chelate reductase
{Pisum sativum}, partial (24%)
Length = 679
Score = 208 bits (530), Expect = 5e-54
Identities = 98/173 (56%), Positives = 131/173 (75%)
Frame = +3
Query: 479 VLGPNNWLWLGAIITSSFVMFLLFLGIVTRYYIYPIENNSGEVYNWTYGVMWYMFSFCSC 538
+LGPN+WLWLGAII+SSF++FL+ +G++TRYYI+PI++N+ +++ M C
Sbjct: 9 MLGPNSWLWLGAIISSSFIIFLIIIGVITRYYIFPIDHNTNAIFSDPLRAFLNMLVICVS 188
Query: 539 ICICSSVVFLWLKRLNKLENKHIMNVEVSTPARSPGSWIYGSERELESLPHQSLVQATNV 598
I + SSV LW K+ N E K I N+E S+P SP S IY ++RELESLP+QSLV+ATNV
Sbjct: 189 IAVVSSVAVLWNKQ-NAKEAKQIQNLEGSSPTVSPSSMIYNADRELESLPYQSLVEATNV 365
Query: 599 HFGSRPDLKKILFECEGKDVGVMTCGPRKMRHEVARICASGLADNLHFESISF 651
H+G RPDLK++LFE +G VGV+ GP++MR EVA IC+SGL +NLHFESISF
Sbjct: 366 HYGQRPDLKRLLFEMKGSSVGVLVSGPKQMRQEVASICSSGLVENLHFESISF 524
>TC90122
Length = 810
Score = 148 bits (374), Expect(2) = 1e-35
Identities = 70/75 (93%), Positives = 71/75 (94%)
Frame = +1
Query: 1 MKRHTIIFLRIIFLVMFLGWLSVWILLPTKIYKKIWAPMLQNKFNSTYFRGQGNYVFVFT 60
MKRHTIIFLRIIFLVMFLGWLSVWILLPTKIYKKIWAPMLQNKFNSTYFRGQG + FT
Sbjct: 490 MKRHTIIFLRIIFLVMFLGWLSVWILLPTKIYKKIWAPMLQNKFNSTYFRGQGINLLFFT 669
Query: 61 FPMMFIGALSCIYLH 75
FPMMFIGALSCIYLH
Sbjct: 670 FPMMFIGALSCIYLH 714
Score = 20.4 bits (41), Expect(2) = 1e-35
Identities = 8/9 (88%), Positives = 9/9 (99%)
Frame = +2
Query: 76 AS*DVHSWL 84
AS*+VHSWL
Sbjct: 782 AS*NVHSWL 808
>BG644856 similar to GP|18087544|gb| AT5g23980/MZF18_14 {Arabidopsis
thaliana}, partial (12%)
Length = 582
Score = 104 bits (259), Expect(2) = 6e-33
Identities = 63/143 (44%), Positives = 78/143 (54%), Gaps = 23/143 (16%)
Frame = +1
Query: 16 MFLGWLSVWILLPTKIYKKIWAPMLQNKFNSTYFRGQGNYVFVFTFPMMFIGALSCIYL- 74
MFLGWL+VWILLPTK K P ++ +G +F F+ + L +Y+
Sbjct: 1 MFLGWLTVWILLPTKSTKTHGLPS*KSSSILHTLESKG*-IFCFSHSLSCSLELWVVYIS 177
Query: 75 ----------------------HAS*DVHSWLCLQLELFQLWSLFLYSCLLHS*YGPFPI 112
+ S*DVHSW CLQLELFQLWSLFL SCLLH *YGPF I
Sbjct: 178 IFIVRKQHKSYPPQVLAL*KNVYVS*DVHSW*CLQLELFQLWSLFLQSCLLHF*YGPFTI 357
Query: 113 TYIPVSITSACTNKMRKFTRMSS 135
T + V +TS CT K +K+ + SS
Sbjct: 358 T*VSVLLTSICTKKAKKYGKRSS 426
Score = 55.5 bits (132), Expect(2) = 6e-33
Identities = 28/30 (93%), Positives = 28/30 (93%)
Frame = +3
Query: 131 TRMSSILPLVGLTSESSIKYHIWLGQLSMV 160
TR SSIL LVGLTSESSIKYHIWLGQLSMV
Sbjct: 492 TRGSSILRLVGLTSESSIKYHIWLGQLSMV 581
>TC78637 similar to GP|8978270|dbj|BAA98161.1 FRO2-like protein; NADPH
oxidase-like {Arabidopsis thaliana}, partial (29%)
Length = 1190
Score = 82.8 bits (203), Expect(2) = 5e-26
Identities = 37/77 (48%), Positives = 55/77 (71%)
Frame = +2
Query: 191 SNVAGEIASLIALAMWITSIPQIRRKMYEVFFYTHHLYILYILFFAIHAGVGSMCVIAPG 250
+N+AG I+ + L MW+TS+P +R +E+FFYTH LYI++I+F A+H G + A
Sbjct: 956 ANLAGVISLVAGLLMWVTSLPGVRTWNFELFFYTHQLYIIFIVFMALHIGDFIFAMAAGP 1135
Query: 251 VFLFLIDRHLRFLQSRQ 267
+FLF++DR LRF QSR+
Sbjct: 1136 IFLFVLDRFLRFCQSRK 1186
Score = 53.5 bits (127), Expect(2) = 5e-26
Identities = 22/58 (37%), Positives = 37/58 (62%), Gaps = 1/58 (1%)
Frame = +1
Query: 131 TRMSSILPLVGLTSESSIKYHIWLGQLSMVLFAAHSIGFIIYWAITNQMI-EMLEWSK 187
+R S +L + + E + KYH+WLG L+MV+F H + ++I W + +I E+LEW +
Sbjct: 772 SRGSVLLRFIDIPFEHAAKYHVWLGHLTMVIFTLHGLFYVIEWLMEGHLIQELLEWKR 945
>CA921657 similar to PIR|T03826|T03 cytochrome b245 beta chain homolog
RbohAp108 - Arabidopsis thaliana, partial (26%)
Length = 749
Score = 94.7 bits (234), Expect = 1e-19
Identities = 63/217 (29%), Positives = 103/217 (47%), Gaps = 32/217 (14%)
Frame = -2
Query: 211 PQIRRKMYEVFFYTHHLYILYILFFAIHAGVGSMCV-----------IAPGVFLFLIDRH 259
P R + F+Y+HHL+++ + IH GV V + + L+ +R
Sbjct: 721 PFSRLTGFNAFWYSHHLFVIVYVLLIIH-GVKLYLVHKWYYQTTWMYLTVPLLLYASERT 545
Query: 260 LRFLQSRQHA-RLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHPFTVT 318
LR +S + RL+ + P + L L SK P Y +F+ VS +WHPF++T
Sbjct: 544 LRLFRSGLYTVRLIKVAIYPGNVLTLQMSKPPQFRYKSGQYMFVQCAAVSPFEWHPFSIT 365
Query: 319 SSCNMETNYLSVAIKNVGSWSNKLYQ--------------------ELSSSSLDHLNISV 358
S+ +YLSV I+ +G W+ +L + E + SL L I
Sbjct: 364 SAPG--DDYLSVHIRQLGDWTQELKRVFSEVCEPPVSGRSGLLRADETTKKSLPKLKI-- 197
Query: 359 EGPYGPHTAQFLRHEQIVMVSGGSGVTPFISIIRDLI 395
+GPYG + +++ +++V G G TPFISI++DL+
Sbjct: 196 DGPYGAPAQDYRKYDVLLLVGLGIGATPFISILKDLL 86
>TC80651 similar to GP|15341529|gb|AAK95654.1 ferric-chelate reductase
{Pisum sativum}, partial (32%)
Length = 984
Score = 72.8 bits (177), Expect = 4e-13
Identities = 27/75 (36%), Positives = 47/75 (62%)
Frame = +3
Query: 1 MKRHTIIFLRIIFLVMFLGWLSVWILLPTKIYKKIWAPMLQNKFNSTYFRGQGNYVFVFT 60
M R +R++ +++FLG + +WI++PT + W P ++ K + TYF QG + ++T
Sbjct: 222 MAREAQSIIRLLVVLLFLGCIMIWIMMPTNTFNLKWFPKIRGKADPTYFGAQGETILMYT 401
Query: 61 FPMMFIGALSCIYLH 75
FP++ I L C+YLH
Sbjct: 402 FPVLLIATLGCVYLH 446
Score = 50.8 bits (120), Expect(2) = 4e-15
Identities = 24/42 (57%), Positives = 29/42 (68%)
Frame = +1
Query: 162 FAAHSIGFIIYWAITNQMIEMLEWSKTYVSNVAGEIASLIAL 203
F H I +I YWA TN+M EML W+K VSN+AGEI+ L L
Sbjct: 856 FTTHGICYITYWASTNKMSEMLIWTKDGVSNLAGEISLLAGL 981
Score = 48.9 bits (115), Expect(2) = 4e-15
Identities = 21/32 (65%), Positives = 25/32 (77%)
Frame = +3
Query: 131 TRMSSILPLVGLTSESSIKYHIWLGQLSMVLF 162
TR SS+LP+ GLTSE IKYHIWLG + M +F
Sbjct: 762 TRGSSVLPIFGLTSEGCIKYHIWLGHILMAIF 857
>BG452927 weakly similar to GP|20268772|gb putative FRO2; NADPH oxidase
{Arabidopsis thaliana}, partial (17%)
Length = 610
Score = 79.0 bits (193), Expect = 5e-15
Identities = 50/202 (24%), Positives = 103/202 (50%), Gaps = 12/202 (5%)
Frame = +1
Query: 317 VTSSCNMETNYLSVAIKNVGSWSNKLYQELSSSSLDH---------LNISVEGPYGPHTA 367
V+SS N+++V IK +G W+ L + ++ + SVEGPYG
Sbjct: 1 VSSSPLDGKNHIAVLIKVLGKWTGGLRERITDGDATEDLSVPPHMVVTASVEGPYGHEVP 180
Query: 368 QFLRHEQIVMVSGGSGVTPFISIIRDLIFQSQQ-QEFQPPKLLLVCIFKNYADLTMLDLM 426
L +E +++V+GG G++PF++I+ D++ + ++ + +P +L+V K +L +L +
Sbjct: 181 YHLMYENLILVAGGIGLSPFLAILSDILHRVREGKPCRPRNILIVWAVKKSNELPLLSTV 360
Query: 427 LPISGLKTRISQLQLQIEAYITREKQEPSKD--TQKQIQTIWFKTNLSDCPISAVLGPNN 484
+ ++ + + ++TRE P ++ K I+++ SD +S ++G N
Sbjct: 361 DMETICPCFSDKVNINVHIFVTRESDPPLEEGYNYKPIKSLCPFPMPSDYGMSGLVGTGN 540
Query: 485 WLWLGAIITSSFVMFLLFLGIV 506
W G + SS + F++ L ++
Sbjct: 541 NFWSGLYVISSTLGFVILLALL 606
>TC77318 similar to GP|16549089|dbj|BAB70751. respiratory burst oxidase
homolog {Solanum tuberosum}, partial (38%)
Length = 1394
Score = 78.6 bits (192), Expect = 7e-15
Identities = 51/178 (28%), Positives = 87/178 (48%), Gaps = 22/178 (12%)
Frame = +2
Query: 247 IAPGVFLFLIDRHLR-FLQSRQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVP 305
+A + ++ +R LR F + ++L + P + L L+ SK Y+ +F+N
Sbjct: 98 LAIPMIIYACERLLRAFRSGYKSVKILKVAVYPGNVLALHVSKPQGFKYHSGQYIFVNCA 277
Query: 306 KVSKLQWHPFTVTSSCNMETNYLSVAIKNVGSWSNKLYQE---------------LSSSS 350
VS QWHPF++TS+ +Y+SV I+ G W+++L L +
Sbjct: 278 DVSPFQWHPFSITSAPG--DDYVSVHIRTAGDWTSQLKAVFAKACQPASGDQSGLLRADM 451
Query: 351 LDHLNIS------VEGPYGPHTAQFLRHEQIVMVSGGSGVTPFISIIRDLIFQSQQQE 402
L NI ++GPYG + +E I++V G G TP ISI++D++ +QQ+
Sbjct: 452 LQGNNIPRMPKLLIDGPYGAPAQDYKDYEVILLVGLGIGATPLISILKDVLNNMKQQK 625
>TC92277 similar to GP|16549089|dbj|BAB70751. respiratory burst oxidase
homolog {Solanum tuberosum}, partial (34%)
Length = 949
Score = 72.0 bits (175), Expect(2) = 5e-14
Identities = 41/156 (26%), Positives = 75/156 (47%), Gaps = 22/156 (14%)
Frame = +3
Query: 262 FLQSRQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHPFTVTSSC 321
F + R+L + P + L L+ SK Y +++N +S +WHPF++TS+
Sbjct: 483 FRSGYKSVRILKVAVYPGNVLALHASKPQGFKYTSGQYIYVNCSDISPFEWHPFSITSAP 662
Query: 322 NMETNYLSVAIKNVGSWSNKLYQELS---------------------SSSLDHL-NISVE 359
+Y+SV I+ +G W+++L + SSL + + ++
Sbjct: 663 G--DDYISVHIRTLGDWTSQLKGIFAKACQPANDDQSGLLRADMLPGKSSLPRMPRLRID 836
Query: 360 GPYGPHTAQFLRHEQIVMVSGGSGVTPFISIIRDLI 395
GPYG + +E +++V G G TP ISI++D++
Sbjct: 837 GPYGAPAQDYKNYEVLLLVGLGIGATPLISILKDVL 944
Score = 23.9 bits (50), Expect(2) = 5e-14
Identities = 12/46 (26%), Positives = 28/46 (60%), Gaps = 3/46 (6%)
Frame = +1
Query: 209 SIPQIRRKM--YEVFFYTHHLY-ILYILFFAIHAGVGSMCVIAPGV 251
++P+ +K+ + F+Y+HHL+ I+Y+ + H+ + S + G+
Sbjct: 283 NLPKPLKKLTGFNAFWYSHHLFVIVYVPIY--HSWIPSFTLARNGI 414
>TC86547 similar to GP|20805911|gb|AAM28891.1 NADPH oxidase {Nicotiana
tabacum}, partial (57%)
Length = 2214
Score = 71.6 bits (174), Expect = 9e-13
Identities = 44/148 (29%), Positives = 75/148 (49%), Gaps = 13/148 (8%)
Frame = +1
Query: 208 TSIPQIRRKM--YEVFFYTHHLYILYILFFAIHAGV----------GSMCVIAPGVFLFL 255
T +P+ K+ + F+Y+HHL+I+ IH + +A + ++
Sbjct: 1591 TKLPKPFNKLTGFNAFWYSHHLFIIVYAMLIIHGTKLYLTKEWNHKTTWMYLAIPITIYG 1770
Query: 256 IDRHLRFLQSR-QHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHP 314
++R +R L+S + R+L + P + L +N SK Y + +N VS L+WHP
Sbjct: 1771 LERLIRALRSSIKSVRILKVAVYPGNVLAINMSKPQGFSYKSGQYMLVNCAAVSPLEWHP 1950
Query: 315 FTVTSSCNMETNYLSVAIKNVGSWSNKL 342
F++TS+ N +YLSV IK +G W+ L
Sbjct: 1951 FSITSAPN--DDYLSVHIKILGDWTRSL 2028
>TC77319 similar to GP|16549089|dbj|BAB70751. respiratory burst oxidase
homolog {Solanum tuberosum}, partial (42%)
Length = 1137
Score = 67.8 bits (164), Expect = 1e-11
Identities = 38/146 (26%), Positives = 73/146 (49%), Gaps = 13/146 (8%)
Frame = +2
Query: 210 IPQIRRKM--YEVFFYTHHLYILYILFFAIHAGV----------GSMCVIAPGVFLFLID 257
+P + +K+ + F+Y+HHL+++ +H + +A + ++ +
Sbjct: 581 LPPLLKKLTGFNAFWYSHHLFVIVYALLIVHGYFLYLSKKWYKKTTWMYLAIPMIIYACE 760
Query: 258 RHLR-FLQSRQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHPFT 316
R LR F + ++L + P + L L+ SK Y+ +F+N VS QWHPF+
Sbjct: 761 RLLRAFRSGYKSVKILKVAVYPGNVLALHVSKPQGFKYHSGQYIFVNCADVSPFQWHPFS 940
Query: 317 VTSSCNMETNYLSVAIKNVGSWSNKL 342
+TS+ +Y+SV I+ G W+++L
Sbjct: 941 ITSAPG--DDYVSVHIRTAGDWTSQL 1012
>AW329243 similar to GP|16549089|db respiratory burst oxidase homolog
{Solanum tuberosum}, partial (18%)
Length = 627
Score = 55.5 bits (132), Expect = 6e-08
Identities = 29/97 (29%), Positives = 53/97 (53%), Gaps = 1/97 (1%)
Frame = +3
Query: 247 IAPGVFLFLIDRHLR-FLQSRQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVP 305
+A + L+ +R LR F + R+L + P + L L+ SK Y +++N
Sbjct: 198 LAVPMILYGCERLLRAFRSGYKSVRILKVAVYPGNVLALHASKPQGFKYTSGQYIYVNCS 377
Query: 306 KVSKLQWHPFTVTSSCNMETNYLSVAIKNVGSWSNKL 342
+S +WHPF++TS+ +Y+SV I+ +G W+++L
Sbjct: 378 DISPFEWHPFSITSAPG--DDYISVHIRTLGDWTSQL 482
>AL365804 weakly similar to GP|18377668|gb| putative FRO1 and FRO2 protein
{Arabidopsis thaliana}, partial (11%)
Length = 479
Score = 47.8 bits (112), Expect = 1e-05
Identities = 26/70 (37%), Positives = 42/70 (59%)
Frame = +3
Query: 5 TIIFLRIIFLVMFLGWLSVWILLPTKIYKKIWAPMLQNKFNSTYFRGQGNYVFVFTFPMM 64
TI+ L IIF+ W+S+WIL PT+++ K W + Q+ N+T F G V+TFP++
Sbjct: 33 TILKLMIIFICA--AWISLWILKPTQVWTKKWKHVEQSA-NNTIFGYYGLNFAVYTFPII 203
Query: 65 FIGALSCIYL 74
I + ++L
Sbjct: 204AIAIIGLLFL 233
>AL365805 similar to GP|18377668|gb| putative FRO1 and FRO2 protein
{Arabidopsis thaliana}, partial (3%)
Length = 503
Score = 44.7 bits (104), Expect = 1e-04
Identities = 22/51 (43%), Positives = 31/51 (60%), Gaps = 4/51 (7%)
Frame = +3
Query: 590 QSLVQATNVHFGSRPDLKKIL--FECE--GKDVGVMTCGPRKMRHEVARIC 636
++ ++ VHFG RP+ K IL F+ E G D+GV+ CGP M+ VA C
Sbjct: 69 KNALEEHEVHFGGRPNFKDILGKFKNETCGSDIGVLVCGPESMKESVASAC 221
>AW687772 similar to GP|16549089|db respiratory burst oxidase homolog
{Solanum tuberosum}, partial (13%)
Length = 622
Score = 40.4 bits (93), Expect = 0.002
Identities = 17/45 (37%), Positives = 29/45 (63%)
Frame = +1
Query: 358 VEGPYGPHTAQFLRHEQIVMVSGGSGVTPFISIIRDLIFQSQQQE 402
++GPYG + +E I++V G G TP ISI++D++ +QQ+
Sbjct: 250 IDGPYGAPAQDYKDYEVILLVGLGIGATPLISILKDVLNNMKQQK 384
>TC80702 similar to GP|8978271|dbj|BAA98162.1 FRO1-like protein; NADPH
oxidase-like {Arabidopsis thaliana}, partial (13%)
Length = 777
Score = 40.0 bits (92), Expect = 0.003
Identities = 24/78 (30%), Positives = 37/78 (46%), Gaps = 10/78 (12%)
Frame = +2
Query: 585 ESLPHQSLVQATNVHFGSRPDLKKILFECEGK----DVGVMTCGPRKMRHEVARICASGL 640
+ L ++ + T + +GSRPD K+I K DVG++ CGP ++ VA+ S
Sbjct: 167 KDLTQYNIAKLTTIRYGSRPDFKEIFESMSEKWGHVDVGILVCGPPTLQSSVAQEIRSHS 346
Query: 641 ADN------LHFESISFN 652
HF S SF+
Sbjct: 347 LTRQPYHPIFHFHSHSFD 400
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.329 0.140 0.449
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,050,062
Number of Sequences: 36976
Number of extensions: 464132
Number of successful extensions: 4250
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 4097
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4216
length of query: 652
length of database: 9,014,727
effective HSP length: 102
effective length of query: 550
effective length of database: 5,243,175
effective search space: 2883746250
effective search space used: 2883746250
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC121237.9


