

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121234.2 + phase: 0
(1118 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
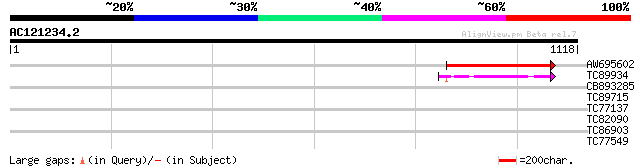
Score E
Sequences producing significant alignments: (bits) Value
AW695602 similar to GP|20147224|g At3g54435 {Arabidopsis thalian... 446 e-125
TC89934 GP|1000353|gb|AAA76715.1| beta-galactosidase {synthetic ... 100 5e-21
CB893285 weakly similar to GP|5091541|gb T10O24.10 {Arabidopsis ... 32 1.8
TC89715 similar to GP|12003384|gb|AAG43549.1 Avr9/Cf-9 rapidly e... 31 2.3
TC77137 similar to PIR|S47140|S47140 pathogenesis-related protei... 30 6.8
TC82090 similar to GP|21387055|gb|AAM47931.1 transmembrane prote... 30 6.8
TC86903 similar to GP|21617881|gb|AAM66931.1 prolyl 4-hydroxylas... 29 8.9
TC77549 similar to GP|21592984|gb|AAM64933.1 photolyase/blue-lig... 29 8.9
>AW695602 similar to GP|20147224|g At3g54435 {Arabidopsis thaliana}, partial
(14%)
Length = 650
Score = 446 bits (1147), Expect = e-125
Identities = 216/216 (100%), Positives = 216/216 (100%)
Frame = +3
Query: 861 SYLSRWKAAGIDSVHFIAESCSVQSTTGNAVKLLVVFHGVTKGEEGSLPNQDKSKVLFTT 920
SYLSRWKAAGIDSVHFIAESCSVQSTTGNAVKLLVVFHGVTKGEEGSLPNQDKSKVLFTT
Sbjct: 3 SYLSRWKAAGIDSVHFIAESCSVQSTTGNAVKLLVVFHGVTKGEEGSLPNQDKSKVLFTT 182
Query: 921 EMTYTIYASGDVILECNVKPNADLPPLPRVGIEMNLEKSLDQVSWYGRGPFECYPDRKAA 980
EMTYTIYASGDVILECNVKPNADLPPLPRVGIEMNLEKSLDQVSWYGRGPFECYPDRKAA
Sbjct: 183 EMTYTIYASGDVILECNVKPNADLPPLPRVGIEMNLEKSLDQVSWYGRGPFECYPDRKAA 362
Query: 981 AQVAVYEKSVDELHVPYIVPGESGGRADVRWATFLNKNGFGIYTSKYGSSPPMQMSASYY 1040
AQVAVYEKSVDELHVPYIVPGESGGRADVRWATFLNKNGFGIYTSKYGSSPPMQMSASYY
Sbjct: 363 AQVAVYEKSVDELHVPYIVPGESGGRADVRWATFLNKNGFGIYTSKYGSSPPMQMSASYY 542
Query: 1041 STSELDRAGHDYELVKGDNIEVHLDHKHMGLGGDDS 1076
STSELDRAGHDYELVKGDNIEVHLDHKHMGLGGDDS
Sbjct: 543 STSELDRAGHDYELVKGDNIEVHLDHKHMGLGGDDS 650
>TC89934 GP|1000353|gb|AAA76715.1| beta-galactosidase {synthetic construct},
partial (25%)
Length = 793
Score = 99.8 bits (247), Expect = 5e-21
Identities = 67/236 (28%), Positives = 110/236 (46%), Gaps = 6/236 (2%)
Frame = +2
Query: 846 FWRASIDNDKGGGA------DSYLSRWKAAGIDSVHFIAESCSVQSTTGNAVKLLVVFHG 899
F RA +DND G ++++ RWKAAG H+ AE+ +Q T +++
Sbjct: 152 FTRAPLDNDIGVSEATRIDPNAWVERWKAAG----HYQAEAALLQCTADTLADAVLITTA 319
Query: 900 VTKGEEGSLPNQDKSKVLFTTEMTYTIYASGDVILECNVKPNADLPPLPRVGIEMNLEKS 959
+G K LF + TY I SG + + +V+ +D P R+G+ L +
Sbjct: 320 HAWQHQG--------KTLFISRKTYRIDGSGQMAITVDVEVASDTPHPARIGLNCQLAQV 475
Query: 960 LDQVSWYGRGPFECYPDRKAAAQVAVYEKSVDELHVPYIVPGESGGRADVRWATFLNKNG 1019
++V+W G GP E YPDR AA ++ + +++ PY+ P E+G R R +
Sbjct: 476 AERVNWLGLGPQENYPDRLTAACFDRWDLPLSDMYTPYVFPSENGLRCGTRELNYGPHQW 655
Query: 1020 FGIYTSKYGSSPPMQMSASYYSTSELDRAGHDYELVKGDNIEVHLDHKHMGLGGDD 1075
G + Q + S YS +L H + L + ++++ HMG+GGDD
Sbjct: 656 RGDF----------QFNISRYSQQQLMETSHRHLLHAEEGTWLNIEGFHMGIGGDD 793
>CB893285 weakly similar to GP|5091541|gb T10O24.10 {Arabidopsis thaliana},
partial (22%)
Length = 821
Score = 31.6 bits (70), Expect = 1.8
Identities = 20/51 (39%), Positives = 29/51 (56%), Gaps = 3/51 (5%)
Frame = +3
Query: 900 VTKGEEGSLPNQDKSKV---LFTTEMTYTIYASGDVILECNVKPNADLPPL 947
+T+ EG LP+ ++ V + T ++T T A +LE NVKP DLP L
Sbjct: 6 LTRYYEGQLPSISENDVDDKVHTPQITVTEAAEKVSLLEENVKPRTDLPHL 158
>TC89715 similar to GP|12003384|gb|AAG43549.1 Avr9/Cf-9 rapidly elicited
protein 111B {Nicotiana tabacum}, partial (49%)
Length = 947
Score = 31.2 bits (69), Expect = 2.3
Identities = 12/33 (36%), Positives = 17/33 (51%)
Frame = -3
Query: 141 PIYTNVTYPFPLDPPFVPTENPTGCYRMDFHLP 173
P ++N P+PL PP P+ +PT H P
Sbjct: 525 PCFSNTPAPYPLPPPVAPSPSPTPPLSTKAHYP 427
>TC77137 similar to PIR|S47140|S47140 pathogenesis-related protein -
chickpea, complete
Length = 897
Score = 29.6 bits (65), Expect = 6.8
Identities = 15/38 (39%), Positives = 21/38 (54%), Gaps = 1/38 (2%)
Frame = -3
Query: 125 STLPVPSNWQLH-GFDRPIYTNVTYPFPLDPPFVPTEN 161
S L +PS+ G P+ +T FP+DPPF P+ N
Sbjct: 631 SPLALPSSTSFSIGLASPLVWYLTVVFPIDPPFGPSTN 518
>TC82090 similar to GP|21387055|gb|AAM47931.1 transmembrane protein-like
protein {Arabidopsis thaliana}, partial (62%)
Length = 829
Score = 29.6 bits (65), Expect = 6.8
Identities = 45/173 (26%), Positives = 65/173 (37%), Gaps = 1/173 (0%)
Frame = +3
Query: 647 DIPNDLNFCLNGLVWPD-RTAHPVLHEVKFLYQPIKVNLSDGKLEIKNTHFFQTTEGLEF 705
+I N++ + +V PD T P L VK + P NL + + F T E +
Sbjct: 213 EIQNNVVVLADYVVVPDDHTRSPTL-AVK-VTSPYGNNLHHKENTTHGSFAFTTQETGNY 386
Query: 706 SWYISADGYKLGSDKLSLPPIKPQSNYVFDWKSGPWYSLWDSSSSEEIFLTITAKLLNST 765
DG G ++S+ DWK G WDS + +E + +L
Sbjct: 387 LACFWVDGGNQGGHEVSVN---------LDWKIGIAAKDWDSVAKKEKIEGVELELRKLE 539
Query: 766 RWVEAGHVVTTAQVQLPAKRDIVPHAINIGSGNLVVETLGDTIKVSQQDVWDI 818
VEA H AK IV N L + +LG I VS +W +
Sbjct: 540 GAVEAIHENLLYLKGREAKMRIVSERTNGRVAWLSIMSLGICIGVSALQLWHL 698
>TC86903 similar to GP|21617881|gb|AAM66931.1 prolyl 4-hydroxylase putative
{Arabidopsis thaliana}, partial (68%)
Length = 1310
Score = 29.3 bits (64), Expect = 8.9
Identities = 19/56 (33%), Positives = 25/56 (43%), Gaps = 4/56 (7%)
Frame = +3
Query: 1057 GDNIEVHLDHKHMGLGGDDSWSPCVHDQYLVPP----VPYSFSVRLSPVTPATSGH 1108
G+ I H + K + D+SWS C H Y V P FS+ L T + S H
Sbjct: 702 GETIFPHAEGK-LSQPKDESWSECAHKGYAVKPRKGDALLFFSLHLDATTDSKSLH 866
>TC77549 similar to GP|21592984|gb|AAM64933.1 photolyase/blue-light receptor
PHR2 {Arabidopsis thaliana}, partial (72%)
Length = 1653
Score = 29.3 bits (64), Expect = 8.9
Identities = 11/21 (52%), Positives = 17/21 (80%), Gaps = 1/21 (4%)
Frame = +3
Query: 143 YTNVTYPFPL-DPPFVPTENP 162
+T+++ PF L PPF+PT+NP
Sbjct: 111 FTSLSLPFSLLSPPFIPTQNP 173
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.136 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 43,307,053
Number of Sequences: 36976
Number of extensions: 717567
Number of successful extensions: 3010
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 2970
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3009
length of query: 1118
length of database: 9,014,727
effective HSP length: 106
effective length of query: 1012
effective length of database: 5,095,271
effective search space: 5156414252
effective search space used: 5156414252
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC121234.2


