

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121234.11 - phase: 0 /pseudo
(92 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
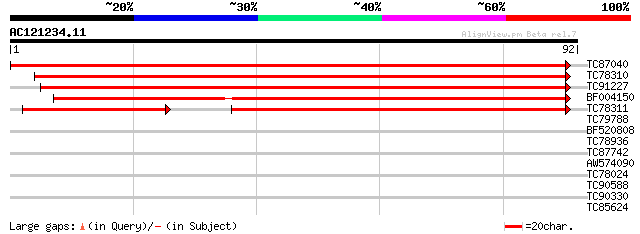
Score E
Sequences producing significant alignments: (bits) Value
TC87040 similar to GP|5002232|gb|AAD37373.1| NADPH:quinone oxido... 178 4e-46
TC78310 similar to GP|11994494|dbj|BAB02535. NADPH:quinone oxido... 134 6e-33
TC91227 similar to GP|11994494|dbj|BAB02535. NADPH:quinone oxido... 126 2e-30
BF004150 similar to GP|11994494|db NADPH:quinone oxidoreductase ... 114 8e-27
TC78311 similar to PIR|B84712|B84712 probable protein kinase [im... 72 4e-14
TC79788 weakly similar to GP|21554189|gb|AAM63268.1 putative leu... 28 0.60
BF520808 similar to GP|11493822|gb| transcription factor WRKY4 {... 28 0.78
TC78936 similar to PIR|A86239|A86239 protein T10O24.17 [imported... 28 0.78
TC87742 weakly similar to GP|7269844|emb|CAB79703.1 serine/threo... 27 1.0
AW574090 26 2.3
TC78024 similar to GP|20804963|dbj|BAB92640. hypothetical protei... 26 3.0
TC90588 similar to SP|P93648|LON2_MAIZE Lon protease homolog 2 ... 25 3.9
TC90330 weakly similar to PIR|D96593|D96593 myosin-like protein ... 24 8.7
TC85624 homologue to SP|P26969|GCSP_PEA Glycine dehydrogenase [d... 24 8.7
>TC87040 similar to GP|5002232|gb|AAD37373.1| NADPH:quinone oxidoreductase
{Arabidopsis thaliana}, partial (94%)
Length = 844
Score = 178 bits (451), Expect = 4e-46
Identities = 91/91 (100%), Positives = 91/91 (100%)
Frame = +3
Query: 1 MATAVIKVAALSGSIRKVSYHSGLIRAAIELSKGATEGIEIEFIDISTLPMYNTDLENEG 60
MATAVIKVAALSGSIRKVSYHSGLIRAAIELSKGATEGIEIEFIDISTLPMYNTDLENEG
Sbjct: 69 MATAVIKVAALSGSIRKVSYHSGLIRAAIELSKGATEGIEIEFIDISTLPMYNTDLENEG 248
Query: 61 TYPPLVEAFRHKILQADSVLFASPEYNYSVT 91
TYPPLVEAFRHKILQADSVLFASPEYNYSVT
Sbjct: 249 TYPPLVEAFRHKILQADSVLFASPEYNYSVT 341
>TC78310 similar to GP|11994494|dbj|BAB02535. NADPH:quinone oxidoreductase
{Arabidopsis thaliana}, partial (92%)
Length = 820
Score = 134 bits (337), Expect = 6e-33
Identities = 65/87 (74%), Positives = 76/87 (86%)
Frame = +2
Query: 5 VIKVAALSGSIRKVSYHSGLIRAAIELSKGATEGIEIEFIDISTLPMYNTDLENEGTYPP 64
VIK+ ALSGS+RK S+H+GLIR AIELSK +GI+IE+ID+S LPM NTDLE EGT+P
Sbjct: 47 VIKIVALSGSLRKASFHTGLIRYAIELSKSVIDGIQIEYIDLSNLPMLNTDLEKEGTFPA 226
Query: 65 LVEAFRHKILQADSVLFASPEYNYSVT 91
VEAFRHKIL+ADSVLFASPEYNYSV+
Sbjct: 227 EVEAFRHKILEADSVLFASPEYNYSVS 307
>TC91227 similar to GP|11994494|dbj|BAB02535. NADPH:quinone oxidoreductase
{Arabidopsis thaliana}, partial (94%)
Length = 857
Score = 126 bits (316), Expect = 2e-30
Identities = 63/86 (73%), Positives = 73/86 (84%)
Frame = +1
Query: 6 IKVAALSGSIRKVSYHSGLIRAAIELSKGATEGIEIEFIDISTLPMYNTDLENEGTYPPL 65
IKV A+SGS+RK SY++GLIR+AIELSK EG++IE+IDISTLPM NTDLE EG +P
Sbjct: 94 IKVVAISGSLRKASYNTGLIRSAIELSKNVVEGVDIEYIDISTLPMLNTDLEIEGNFPAE 273
Query: 66 VEAFRHKILQADSVLFASPEYNYSVT 91
VEAFR KIL ADS LFASPEYNYSV+
Sbjct: 274 VEAFRQKILAADSDLFASPEYNYSVS 351
>BF004150 similar to GP|11994494|db NADPH:quinone oxidoreductase
{Arabidopsis thaliana}, partial (63%)
Length = 379
Score = 114 bits (284), Expect = 8e-27
Identities = 59/84 (70%), Positives = 70/84 (83%)
Frame = +1
Query: 8 VAALSGSIRKVSYHSGLIRAAIELSKGATEGIEIEFIDISTLPMYNTDLENEGTYPPLVE 67
VAALSGS+RK SY++GLIR+AIELSK EG+ IE+++IS LP+ NTDLE EGTYP VE
Sbjct: 1 VAALSGSLRKASYNTGLIRSAIELSKNV-EGVHIEYVNISNLPLLNTDLEKEGTYPVEVE 177
Query: 68 AFRHKILQADSVLFASPEYNYSVT 91
+FR KIL ADS LFASPE NYSV+
Sbjct: 178SFRKKILGADSFLFASPENNYSVS 249
>TC78311 similar to PIR|B84712|B84712 probable protein kinase [imported] -
Arabidopsis thaliana, partial (44%)
Length = 1323
Score = 72.0 bits (175), Expect = 4e-14
Identities = 34/55 (61%), Positives = 42/55 (75%)
Frame = +3
Query: 37 EGIEIEFIDISTLPMYNTDLENEGTYPPLVEAFRHKILQADSVLFASPEYNYSVT 91
+ +E ++DIS LP+ NTDLE EGT+P VE FR IL ADS LFASPEYNYS++
Sbjct: 279 DDMEFVYVDISNLPLLNTDLEEEGTFPLEVEDFRDYILGADSFLFASPEYNYSIS 443
Score = 38.1 bits (87), Expect = 6e-04
Identities = 17/24 (70%), Positives = 21/24 (86%)
Frame = +2
Query: 3 TAVIKVAALSGSIRKVSYHSGLIR 26
T VIK+A LSGS+RK S+H+GLIR
Sbjct: 8 TGVIKIACLSGSLRKGSFHTGLIR 79
>TC79788 weakly similar to GP|21554189|gb|AAM63268.1 putative leucine-rich
repeat disease resistance protein {Arabidopsis
thaliana}, partial (52%)
Length = 1484
Score = 28.1 bits (61), Expect = 0.60
Identities = 17/49 (34%), Positives = 27/49 (54%), Gaps = 1/49 (2%)
Frame = +3
Query: 6 IKVAALSGSIRKVSYHSGLIRAAIELSKGATEG-IEIEFIDISTLPMYN 53
IK +LSG ++K ++ +ELS+ A G IE F+ +S+L N
Sbjct: 687 IKANSLSGPLQKTTFEGSNQLEVVELSENALTGTIETWFLLLSSLQQVN 833
>BF520808 similar to GP|11493822|gb| transcription factor WRKY4 {Petroselinum
crispum}, partial (16%)
Length = 585
Score = 27.7 bits (60), Expect = 0.78
Identities = 13/16 (81%), Positives = 13/16 (81%)
Frame = +3
Query: 74 LQADSVLFASPEYNYS 89
L ADSVLFASPEY S
Sbjct: 180 LAADSVLFASPEYKCS 227
>TC78936 similar to PIR|A86239|A86239 protein T10O24.17 [imported] -
Arabidopsis thaliana, partial (72%)
Length = 1325
Score = 27.7 bits (60), Expect = 0.78
Identities = 14/43 (32%), Positives = 22/43 (50%)
Frame = +1
Query: 1 MATAVIKVAALSGSIRKVSYHSGLIRAAIELSKGATEGIEIEF 43
MAT + + SG + + Y G AAI+L G + G+ + F
Sbjct: 223 MATLALDKISGSGLVSQSRYSYGFFSAAIKLPAGLSPGVVVAF 351
>TC87742 weakly similar to GP|7269844|emb|CAB79703.1
serine/threonine-specific receptor protein kinase-like
protein {Arabidopsis thaliana}, partial (29%)
Length = 1318
Score = 27.3 bits (59), Expect = 1.0
Identities = 13/36 (36%), Positives = 20/36 (55%)
Frame = +1
Query: 33 KGATEGIEIEFIDISTLPMYNTDLENEGTYPPLVEA 68
K A+EG+ +E I ++T N L N+G P + A
Sbjct: 577 KNASEGVTLEIISMATSEETNVCLVNKGKGIPFISA 684
>AW574090
Length = 401
Score = 26.2 bits (56), Expect = 2.3
Identities = 17/44 (38%), Positives = 20/44 (44%), Gaps = 4/44 (9%)
Frame = +1
Query: 52 YNTDLE----NEGTYPPLVEAFRHKILQADSVLFASPEYNYSVT 91
YNT LE NE TY P + + + L S L Y SVT
Sbjct: 82 YNTPLEVKESNETTYSPNISSSSFEPLSDFSELLVQTSYQVSVT 213
>TC78024 similar to GP|20804963|dbj|BAB92640. hypothetical protein~similar
to Arabidopsis thaliana chromosome 3 At3g51270, partial
(43%)
Length = 1107
Score = 25.8 bits (55), Expect = 3.0
Identities = 19/67 (28%), Positives = 29/67 (42%)
Frame = +2
Query: 15 IRKVSYHSGLIRAAIELSKGATEGIEIEFIDISTLPMYNTDLENEGTYPPLVEAFRHKIL 74
+R +S + A+EL E + E ID L++ GTY L +HK+L
Sbjct: 155 LRYLSKDDFRVLTAVELGMRNHEIVPAELIDRIAR------LKHGGTYKVLKNLLKHKLL 316
Query: 75 QADSVLF 81
DS +
Sbjct: 317 HHDSTKY 337
>TC90588 similar to SP|P93648|LON2_MAIZE Lon protease homolog 2 mitochondrial
precursor (EC 3.4.21.-). [Maize] {Zea mays}, partial
(34%)
Length = 1540
Score = 25.4 bits (54), Expect = 3.9
Identities = 11/38 (28%), Positives = 20/38 (51%)
Frame = +1
Query: 8 VAALSGSIRKVSYHSGLIRAAIELSKGATEGIEIEFID 45
+AA ++ + + S R EL+ EG+E+ F+D
Sbjct: 1144 IAARRSEVKTIIFPSANKRDFDELAPNVKEGLEVHFVD 1257
>TC90330 weakly similar to PIR|D96593|D96593 myosin-like protein
97843-94399 [imported] - Arabidopsis thaliana, partial
(36%)
Length = 1191
Score = 24.3 bits (51), Expect = 8.7
Identities = 9/24 (37%), Positives = 15/24 (62%)
Frame = -3
Query: 53 NTDLENEGTYPPLVEAFRHKILQA 76
++D ++ YPP +F+H LQA
Sbjct: 259 SSDHQSTSCYPPPASSFQHAALQA 188
>TC85624 homologue to SP|P26969|GCSP_PEA Glycine dehydrogenase
[decarboxylating] mitochondrial precursor (EC 1.4.4.2),
partial (98%)
Length = 3772
Score = 24.3 bits (51), Expect = 8.7
Identities = 13/35 (37%), Positives = 20/35 (57%), Gaps = 3/35 (8%)
Frame = +2
Query: 30 ELSKGATEGI---EIEFIDISTLPMYNTDLENEGT 61
E+S+G E + + D++ LPM N L +EGT
Sbjct: 755 EISQGRLESLLNFQTLITDLTGLPMSNASLLDEGT 859
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.133 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,038,216
Number of Sequences: 36976
Number of extensions: 18537
Number of successful extensions: 102
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 84
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 101
length of query: 92
length of database: 9,014,727
effective HSP length: 68
effective length of query: 24
effective length of database: 6,500,359
effective search space: 156008616
effective search space used: 156008616
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 51 (24.3 bits)
Medicago: description of AC121234.11


