

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC119408.8 + phase: 0
(908 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
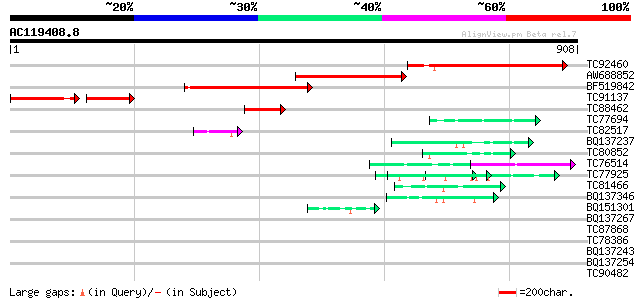
Score E
Sequences producing significant alignments: (bits) Value
TC92460 similar to GP|22202705|dbj|BAC07363. hypothetical protei... 472 e-133
AW688852 weakly similar to PIR|E71443|E714 probable DNA-binding ... 392 e-109
BF519842 similar to GP|22136022|gb| DNA-binding protein-like {Ar... 318 6e-87
TC91137 similar to PIR|D71443|D71443 hypothetical protein - Arab... 172 1e-73
TC88462 55 9e-08
TC77694 homologue to PIR|T09704|T09704 probable arginine/serine-... 49 7e-06
TC82517 homologue to GP|7688065|emb|CAB89694.1 constitutively ph... 49 7e-06
BQ137237 weakly similar to GP|11993889|gb| virion-associated nuc... 47 3e-05
TC80852 similar to GP|14532720|gb|AAK64161.1 unknown protein {Ar... 47 3e-05
TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - ... 46 7e-05
TC77925 similar to GP|18700163|gb|AAL77693.1 AT4g02720/T10P11_1 ... 45 1e-04
TC81466 similar to GP|5821143|dbj|BAA83713.1 RNA binding protein... 44 3e-04
BQ137346 homologue to GP|15027095|em hypothetical protein LT.09 ... 42 8e-04
BQ151301 similar to PIR|S34666|S346 glycine-rich protein - commo... 42 8e-04
BQ137267 similar to GP|9800294|gb|A pR79 {rat cytomegalovirus Ma... 42 0.001
TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 42 0.001
TC78386 similar to SP|Q9NZW4|DSPP_HUMAN Dentin sialophosphoprote... 42 0.001
BQ137243 similar to GP|2624970|gb|A proline-rich protein 9-1 {Mu... 42 0.001
BQ137254 similar to GP|22946423|gb| CG31813-PA {Drosophila melan... 41 0.002
TC90482 41 0.002
>TC92460 similar to GP|22202705|dbj|BAC07363. hypothetical protein {Oryza
sativa (japonica cultivar-group)}, partial (9%)
Length = 795
Score = 472 bits (1215), Expect = e-133
Identities = 244/263 (92%), Positives = 245/263 (92%), Gaps = 7/263 (2%)
Frame = +3
Query: 638 FGREVSSVGDVSSIKSKTKPIPPSSASDYHQNRHRSERPSP-------DSSHREVEPPRP 690
FGREVSSVGDVSSIKSKTKPIPPSSAS S +PSP DSSHREVEPPRP
Sbjct: 3 FGREVSSVGDVSSIKSKTKPIPPSSAS------RLSSKPSPVQNGHRQDSSHREVEPPRP 164
Query: 691 TKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDRHQHRRHHHRTEASSKKSTDPVTKSTS 750
TKRKSDHSEREREDRDRDYDYHDRHQDRDY HDRHQHRRHHHRTEASSKKSTDPVTKSTS
Sbjct: 165 TKRKSDHSEREREDRDRDYDYHDRHQDRDYEHDRHQHRRHHHRTEASSKKSTDPVTKSTS 344
Query: 751 RKSSEPDIKSKSRKSSEPVTKSLSLSKTAPTTSAEAAAAAAADRKQKASVFSRISFPSEE 810
RKSSEPDIKSKSRKSSEPVTKSLSLSKTA TTSAEAAAAAAADRKQKASVFSRISFPSEE
Sbjct: 345 RKSSEPDIKSKSRKSSEPVTKSLSLSKTAQTTSAEAAAAAAADRKQKASVFSRISFPSEE 524
Query: 811 EAAAAAKKRKLSASSTTEASTAATASVSAKAPSTTHLSNGGRKSKAVMDDYESSDDDEDD 870
EAAAAAKKRKLSASSTTEASTAATASVSAKAPSTTHLSNGGRKSKAVMDDYESSDDDEDD
Sbjct: 525 EAAAAAKKRKLSASSTTEASTAATASVSAKAPSTTHLSNGGRKSKAVMDDYESSDDDEDD 704
Query: 871 ERHFKRRPSRYEPSPPPPVDDWV 893
ERHFKRRPSRYEPSPPPPVDDWV
Sbjct: 705 ERHFKRRPSRYEPSPPPPVDDWV 773
>AW688852 weakly similar to PIR|E71443|E714 probable DNA-binding protein -
Arabidopsis thaliana, partial (9%)
Length = 536
Score = 392 bits (1008), Expect = e-109
Identities = 178/178 (100%), Positives = 178/178 (100%)
Frame = +2
Query: 458 EPVSQGSAQVVEEEVQQKLVPTEAGKKKKKKKVRMPTNDFQWKPPHDLGAENYMMQMGPP 517
EPVSQGSAQVVEEEVQQKLVPTEAGKKKKKKKVRMPTNDFQWKPPHDLGAENYMMQMGPP
Sbjct: 2 EPVSQGSAQVVEEEVQQKLVPTEAGKKKKKKKVRMPTNDFQWKPPHDLGAENYMMQMGPP 181
Query: 518 PGYNPYWNGMQPCMDGFMAPYAGPMHMMDYGHGPYDMPFPNGMPHDPFANGMPHDPFGMQ 577
PGYNPYWNGMQPCMDGFMAPYAGPMHMMDYGHGPYDMPFPNGMPHDPFANGMPHDPFGMQ
Sbjct: 182 PGYNPYWNGMQPCMDGFMAPYAGPMHMMDYGHGPYDMPFPNGMPHDPFANGMPHDPFGMQ 361
Query: 578 GYMMPPIPPPPHRDLAEFSMGMNVPPPAMSREEFEARKADARRKRENERRVERDFSKD 635
GYMMPPIPPPPHRDLAEFSMGMNVPPPAMSREEFEARKADARRKRENERRVERDFSKD
Sbjct: 362 GYMMPPIPPPPHRDLAEFSMGMNVPPPAMSREEFEARKADARRKRENERRVERDFSKD 535
>BF519842 similar to GP|22136022|gb| DNA-binding protein-like {Arabidopsis
thaliana}, partial (19%)
Length = 623
Score = 318 bits (815), Expect = 6e-87
Identities = 163/208 (78%), Positives = 178/208 (85%), Gaps = 2/208 (0%)
Frame = +3
Query: 280 EKEMEGMPSTTRSVGDLPPELHCPLCSNVMKDAVLTSKCCFKSFCDKCIRDYIISKSMCV 339
+KEMEG+ S+TRSVGDLPPELHCPLC+NVMKDAVLTSKCCFKSFCDKCIR+YI+SKS CV
Sbjct: 3 DKEMEGL-SSTRSVGDLPPELHCPLCNNVMKDAVLTSKCCFKSFCDKCIRNYIMSKSACV 179
Query: 340 CGAMNVLADDLLPNKTLRDTINRILESGNSSTENAGSTYQVQDMESARCPQPKIPSPTSS 399
C A N+LADDLLPNKTLRD I+RILESGNSSTENAGSTYQVQDM S+RCPQPKIPSPTSS
Sbjct: 180 CLATNILADDLLPNKTLRDAIHRILESGNSSTENAGSTYQVQDMLSSRCPQPKIPSPTSS 359
Query: 400 AASKGGLKISPVYDGTTNIQDTAVETKVVSAPPQTSEHVKIPRAGDVSEATHESKSVK-- 457
A SKG K+S V +G NIQ+ E K VSA Q SE VKIPRA VSE THES SVK
Sbjct: 360 ATSKGEPKVSQVNEGMANIQEIVAERKEVSATQQVSEQVKIPRAAVVSEVTHESMSVKEP 539
Query: 458 EPVSQGSAQVVEEEVQQKLVPTEAGKKK 485
EP SQGSA++VEEEVQQKLVPT+ GKKK
Sbjct: 540 EPASQGSAKLVEEEVQQKLVPTDRGKKK 623
>TC91137 similar to PIR|D71443|D71443 hypothetical protein - Arabidopsis
thaliana, partial (41%)
Length = 812
Score = 172 bits (435), Expect(2) = 1e-73
Identities = 88/112 (78%), Positives = 97/112 (86%)
Frame = +1
Query: 1 MAVYYKFKSARDYDSISMDGPFISVGTLKEKIFESKHLGRGTDFDLVVTNAQTNEEYLDE 60
MAVYYKFKSA+DY+SI MDGPFISVGTLK+ IF SKHLGRGTDFDL+VTNAQTNEEY DE
Sbjct: 235 MAVYYKFKSAKDYNSIPMDGPFISVGTLKQSIFVSKHLGRGTDFDLLVTNAQTNEEYHDE 414
Query: 61 EMLIPKNTSVLIRRVPGRPRLPIVTEQEYYLKPLISQKVENKVVDTEPANSS 112
EMLIPKNTSVLIRRVPGRP LPIVTE++ Q+ ENKVV+TEP NS+
Sbjct: 415 EMLIPKNTSVLIRRVPGRPCLPIVTEKK--------QEFENKVVETEPENST 546
Score = 124 bits (311), Expect(2) = 1e-73
Identities = 60/77 (77%), Positives = 67/77 (86%), Gaps = 1/77 (1%)
Frame = +3
Query: 124 VEDSDWDEFGNDLYSIPDQLPVQSINMIQEAPPTSTVDEESKIKALIDTPALDWQHQGSD 183
+++ DWDEFGNDLYSIPDQLPVQS MI +AP TS DE+SKIKALIDTPALDWQ QGSD
Sbjct: 582 LKNLDWDEFGNDLYSIPDQLPVQSSTMIPDAPLTSKADEDSKIKALIDTPALDWQQQGSD 761
Query: 184 FGAGRGFGRG-MGGRMG 199
FGAGRGF RG +GGR+G
Sbjct: 762 FGAGRGFRRGAVGGRIG 812
>TC88462
Length = 1157
Score = 55.5 bits (132), Expect = 9e-08
Identities = 32/68 (47%), Positives = 43/68 (63%), Gaps = 2/68 (2%)
Frame = -3
Query: 376 STYQVQDMESARCPQPKIPSPTSSAASKGGLKISPVYDGTTNIQDTAVETKVVSAPPQTS 435
S+ +++ MES+ CPQ K+P PT SA SKG K+S V DG NIQ+ A E K+V P +
Sbjct: 735 SS*RIRYMESSCCPQSKLPFPTLSATSKGEPKVSLVNDGMMNIQEIADERKLVCYPTVAA 556
Query: 436 EH--VKIP 441
+KIP
Sbjct: 555 AG*VIKIP 532
>TC77694 homologue to PIR|T09704|T09704 probable arginine/serine-rich
splicing factor - alfalfa, partial (95%)
Length = 1494
Score = 49.3 bits (116), Expect = 7e-06
Identities = 48/181 (26%), Positives = 69/181 (37%), Gaps = 2/181 (1%)
Frame = +3
Query: 672 RSERPSPDSSHREVEPPRPTKRKSDHSERE--REDRDRDYDYHDRHQDRDYXHDRHQHRR 729
RS PS S HR+ D+ E++ R R R YD H+R DR DR RR
Sbjct: 387 RSRSPSKRSRHRD-----------DYKEKDYRRRSRSRSYDRHER--DRHRGRDRDHRRR 527
Query: 730 HHHRTEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTKSLSLSKTAPTTSAEAAAA 789
R+ + K R+S P S+S S PV +S S+ K +P+ +
Sbjct: 528 SRSRSASPGYKGRGRGRHDDERRSRSP---SRSVDSRSPVRRS-SIPKRSPSPKRSPSPK 695
Query: 790 AAADRKQKASVFSRISFPSEEEAAAAAKKRKLSASSTTEASTAATASVSAKAPSTTHLSN 849
+ K+ S +S P + + R S T S + A + SN
Sbjct: 696 RSPSLKRSISPQKSVS-PRKSPLRESPDNRSRGGRSLTPRSVSPRGRPGASRSPSPRNSN 872
Query: 850 G 850
G
Sbjct: 873 G 875
>TC82517 homologue to GP|7688065|emb|CAB89694.1 constitutively
photomorphogenic 1 protein {Pisum sativum}, partial
(22%)
Length = 671
Score = 49.3 bits (116), Expect = 7e-06
Identities = 29/83 (34%), Positives = 43/83 (50%), Gaps = 4/83 (4%)
Frame = +3
Query: 295 DLPPELHCPLCSNVMKDAVLTSKCCFKSFCDKCIRDYIISKSMCVCGAMNVLADDLLPN- 353
+L + CP+C ++KDA LTS C SFC CI ++ +KS C C + +L PN
Sbjct: 138 ELDKDFLCPICMQIIKDAFLTS--CGHSFCYMCIITHLRNKSDCPCCGHYLTNSNLFPNF 311
Query: 354 ---KTLRDTINRILESGNSSTEN 373
K L+ T +R + S E+
Sbjct: 312 LLDKLLKKTSDRQISKTASPVEH 380
>BQ137237 weakly similar to GP|11993889|gb| virion-associated
nuclear-shuttling protein {Mus musculus}, partial (4%)
Length = 1215
Score = 47.4 bits (111), Expect = 3e-05
Identities = 51/246 (20%), Positives = 85/246 (33%), Gaps = 18/246 (7%)
Frame = +3
Query: 612 EARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQNRH 671
+ RK A+ KR+ E R G+E + +I + P ++ +H R
Sbjct: 414 QGRKRHAQEKRKTEGRRRGHGETTGGAGKEATQNEQTPTITTHEHEQPRTTRQQHHTTRP 593
Query: 672 RSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHD----------RHQDRDYX 721
S + HR PP T R+ ++R+R D D + +D
Sbjct: 594 PSHTHNTAVGHRH-HPPTMTDAAESRKTRKNKERERSSDRRDGRRGAAPRAATRRTKDTT 770
Query: 722 HDR--------HQHRRHHHRTEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTKSL 773
HDR H H +H RTE ++ K + RK E T
Sbjct: 771 HDRTRRCSSHLHHHTQHQQRTEQGDRQK-----------------KERRRKEREGAT--- 890
Query: 774 SLSKTAPTTSAEAAAAAAADRKQKASVFSRISFPSEEEAAAAAKKRKLSASSTTEASTAA 833
+++ TA T + +A+ ++R + E KR ++ ++T
Sbjct: 891 TIAHTATRTEHRHRIRRKLQSRDQATQYTRKE--TTERIGGECSKRVRDSTRRKRSATEH 1064
Query: 834 TASVSA 839
TA A
Sbjct: 1065TAKSDA 1082
Score = 29.3 bits (64), Expect = 7.1
Identities = 30/147 (20%), Positives = 62/147 (41%), Gaps = 10/147 (6%)
Frame = +2
Query: 720 YXHDRHQHRRHHH-RTEASSKKSTDPVTKST-SRKSSEPDIKSKSRKSSEP---VTKSLS 774
+ H+R Q + HH ++ + P + T K S+P K+++ + +P T++
Sbjct: 86 FQHEREQKQEHHRGGGRTKTRGAPGPKERGTREEKQSDPTEKARTTRKHKPRPTPTQTQR 265
Query: 775 LSKTAP-----TTSAEAAAAAAADRKQKASVFSRISFPSEEEAAAAAKKRKLSASSTTEA 829
+K AP T+ + A R+++ R P + K R+ + + T EA
Sbjct: 266 RAKGAPKEQGRQTTKKEKTRTRAGREKEGGGAKRTGTPDPGQ-----KDRRATETGTEEA 430
Query: 830 STAATASVSAKAPSTTHLSNGGRKSKA 856
A + + ++GGR+ ++
Sbjct: 431 --RAGEEEDRRKATRARGNDGGRRKRS 505
>TC80852 similar to GP|14532720|gb|AAK64161.1 unknown protein {Arabidopsis
thaliana}, partial (3%)
Length = 902
Score = 47.0 bits (110), Expect = 3e-05
Identities = 41/154 (26%), Positives = 61/154 (38%), Gaps = 6/154 (3%)
Frame = +3
Query: 662 SASDYHQNRH----RSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQD 717
S+S+ ++RH R S D HR R + SDH R R D RH
Sbjct: 45 SSSEEVEHRHLRHQRKYESSSDEKHRSSRRHRGGSKLSDHEHRHSRKRYSSSDDEHRHSR 224
Query: 718 RDY--XHDRHQHRRHHHRTEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTKSLSL 775
+ Y D H+H R H+ S+D SRK+ K KSR +E + L
Sbjct: 225 KRYSSSDDEHRHPRKHY-------SSSDDEHSHRSRKT-----KHKSRSHAEREAE-LEE 365
Query: 776 SKTAPTTSAEAAAAAAADRKQKASVFSRISFPSE 809
+ + ++ + A R+ A +F PS+
Sbjct: 366 GEIVKSDKSQVSEVGRASREASAELFKSTRAPSQ 467
Score = 30.0 bits (66), Expect = 4.2
Identities = 37/152 (24%), Positives = 57/152 (37%), Gaps = 20/152 (13%)
Frame = +3
Query: 712 HDRHQDRDYXHD------RHQHRRHHHRTEASS--KKSTDPVTKSTSRKSSEPDIKSKSR 763
+D++ R+ HD H+H RH + E+SS K + + S+ S S+ R
Sbjct: 9 NDKYSHRESRHDSSSEEVEHRHLRHQRKYESSSDEKHRSSRRHRGGSKLSDHEHRHSRKR 188
Query: 764 KSSEPVTKSLSLSKTAPTT----------SAEAAAAAAADRKQKASVFSRISFPSEEEAA 813
SS S + + + S+ + RK K S +E E
Sbjct: 189 YSSSDDEHRHSRKRYSSSDDEHRHPRKHYSSSDDEHSHRSRKTKHKSRSHAEREAELEEG 368
Query: 814 AAAKKRKLSASSTTEASTAATASV--SAKAPS 843
K K S AS A+A + S +APS
Sbjct: 369 EIVKSDKSQVSEVGRASREASAELFKSTRAPS 464
>TC76514 homologue to PIR|T09648|T09648 nucleolin homolog nuM1 - alfalfa,
partial (61%)
Length = 1288
Score = 45.8 bits (107), Expect = 7e-05
Identities = 43/171 (25%), Positives = 77/171 (44%), Gaps = 3/171 (1%)
Frame = +3
Query: 739 KKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTKSLSLSKTAPTTSAEAAAAAAADRKQKA 798
KK +S+ + +P +K+ + P K TTS E+ + +++ +++
Sbjct: 279 KKEESSSEESSESEDEQPVVKAPAPSKKTPAKKGNVKKAQPETTSEESDSDSSSSDEEEV 458
Query: 799 SVFSRISFPSEEEAAAAAKKRKLSASSTTEASTAATASVSAKAPSTTHLSNGGRKSKAVM 858
+ PS+ +A AKK S +E S+ +AKA + + S +K+ +
Sbjct: 459 KKPVSKAVPSKN-GSAPAKKVDTSEEEDSEESSDEDKKPAAKAVPSKNGSAPAKKAASDE 635
Query: 859 DDY-ESSDDDEDDER-HFKRRPSRYEPSPPPPVD-DWVEEGRHSRGTRDRK 906
+D ESSD+DE+DE+ K PS+ P D + +E S D+K
Sbjct: 636 EDTDESSDEDEEDEKPAAKAVPSKNGSVPAKKADTESSDEDSESSDEEDKK 788
Score = 45.4 bits (106), Expect = 1e-04
Identities = 63/335 (18%), Positives = 128/335 (37%), Gaps = 5/335 (1%)
Frame = +3
Query: 577 QGY-MMPPIPPPPHRDLAEFSMGMNVPPPAMSREEFEARKADARRKRENERRVERDFSKD 635
QG+ +PP PP H L F M + A + + + ++ +R+ E +
Sbjct: 42 QGFEQLPPHPPISHSSLL-FIMPKSSKKSATKVDAAPVVVSPVKSGKKGKRQAEEEV--- 209
Query: 636 RDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQNRHRSERPSPDSSHREVEPPRPTKRKS 695
+ + V +V++ + K + +S + E+P V+ P P+K+
Sbjct: 210 KAVSAKKQKVEEVAAKQKALKVVKKEESSSEESSESEDEQPV-------VKAPAPSKKTP 368
Query: 696 DHSEREREDRDRDYDYHDRHQDRDYXHDRHQHRRHHHRTEASSKKSTDPVTKSTSRKSSE 755
++ + + D D + + SK + P K + S E
Sbjct: 369 AKKGNVKKAQP---ETTSEESDSDSSSSDEEEVKKPVSKAVPSKNGSAPAKKVDT--SEE 533
Query: 756 PDIKSKSRKSSEPVTKSL-SLSKTAPTTSA---EAAAAAAADRKQKASVFSRISFPSEEE 811
D + S + +P K++ S + +AP A E ++D ++ + + PS+
Sbjct: 534 EDSEESSDEDKKPAAKAVPSKNGSAPAKKAASDEEDTDESSDEDEEDEKPAAKAVPSKNG 713
Query: 812 AAAAAKKRKLSASSTTEASTAATASVSAKAPSTTHLSNGGRKSKAVMDDYESSDDDEDDE 871
+ A K S+ +E+S +AKA + ++S +K+ + D+ + DED++
Sbjct: 714 SVPAKKADTESSDEDSESSDEEDKKPAAKA--SKNVSAPTKKAASSSDEESDEESDEDED 887
Query: 872 RHFKRRPSRYEPSPPPPVDDWVEEGRHSRGTRDRK 906
+P+ D +E S D+K
Sbjct: 888 AKPVSKPAAVAKKSKKDSSDSDDEDDDSSSDEDKK 992
Score = 35.0 bits (79), Expect = 0.13
Identities = 25/96 (26%), Positives = 36/96 (37%)
Frame = +3
Query: 391 PKIPSPTSSAASKGGLKISPVYDGTTNIQDTAVETKVVSAPPQTSEHVKIPRAGDVSEAT 450
PK +++ + +SPV G + E K VSA Q E V +
Sbjct: 105 PKSSKKSATKVDAAPVVVSPVKSGKKGKRQAEEEVKAVSAKKQKVEEVAAKQKALKVVKK 284
Query: 451 HESKSVKEPVSQGSAQVVEEEVQQKLVPTEAGKKKK 486
ES S + S+ VV+ K P + G KK
Sbjct: 285 EESSSEESSESEDEQPVVKAPAPSKKTPAKKGNVKK 392
>TC77925 similar to GP|18700163|gb|AAL77693.1 AT4g02720/T10P11_1
{Arabidopsis thaliana}, partial (47%)
Length = 1538
Score = 45.4 bits (106), Expect = 1e-04
Identities = 56/245 (22%), Positives = 95/245 (37%), Gaps = 30/245 (12%)
Frame = +3
Query: 666 YHQNRHRSERPSPDSSHREVEPPRPTKRKSD--------HSEREREDRDRDYDYHDRHQD 717
Y ++RH R +PDS +V RP R+S H++R R + + RH D
Sbjct: 66 YQRHRHNDRRRTPDS---DVSNDRPRYRRSPSYDSYDDRHTDRHRRRSISPENRNPRHHD 236
Query: 718 RDYXHDRHQHRRHHHRTEAS---------SKKSTDPVTKSTSRKSSEPDIKSKSRKS--- 765
+ H ++ HR S ++ +D K S + + K RKS
Sbjct: 237 NGNSNGNHLPKKFGHRNNGSYLDRNDSRRNESESDEELKGLSYEEYRRLKRQKMRKSLKH 416
Query: 766 -------SEPVTKSLSLSKTAPTTSAEAAAAAAADR--KQKASVFSRISFPSEEEAAAAA 816
S P ++ L D+ KQKA S S S+E +
Sbjct: 417 CIWNVTPSPPRRENDDLEDYGKPEEISEIIDVKKDKEIKQKAKSESE-SEESKESDSELR 593
Query: 817 KKRKLSASSTTEASTAATASVSAKAPSTTHLSNGGRKSKAVMD-DYESSDDDEDDERHFK 875
KKR+ S + E+ + + + + RKS+ + D +++ ++ED +R +
Sbjct: 594 KKRRKSYKKSRESDSESESESEVE-------DRKRRKSRKYSESDSDTNSEEEDRKRRRR 752
Query: 876 RRPSR 880
R+ SR
Sbjct: 753 RKSSR 767
Score = 43.5 bits (101), Expect = 4e-04
Identities = 39/165 (23%), Positives = 64/165 (38%), Gaps = 1/165 (0%)
Frame = +3
Query: 586 PPPHRDLAEFSMGMNVPPPAMSREEFEARKADARRKRENERRVERDFSKDRDFGREVSSV 645
P ++ + + A S E E K R+ R+ + K R+ E S
Sbjct: 483 PEEISEIIDVKKDKEIKQKAKSESESEESKESDSELRKKRRK---SYKKSRESDSESESE 653
Query: 646 GDVSSIKS-KTKPIPPSSASDYHQNRHRSERPSPDSSHREVEPPRPTKRKSDHSERERED 704
+V K K++ S + + R R SS R + R +R SD E E +D
Sbjct: 654 SEVEDRKRRKSRKYSESDSDTNSEEEDRKRRRRRKSSRRRRKSSRKKRRCSDSDESETDD 833
Query: 705 RDRDYDYHDRHQDRDYXHDRHQHRRHHHRTEASSKKSTDPVTKST 749
+ YD D + R+ R+ S KKS++PV++ +
Sbjct: 834 -ESGYD------------DSGRKRKRSKRSRKSKKKSSEPVSEGS 929
Score = 42.7 bits (99), Expect = 6e-04
Identities = 52/190 (27%), Positives = 75/190 (39%), Gaps = 24/190 (12%)
Frame = +3
Query: 606 MSREEFEARKADARRKR---------ENERRVERDFSKDRDFGREVSSVGDVSS---IKS 653
+S EE+ K RK + R E D +D E+S + DV IK
Sbjct: 357 LSYEEYRRLKRQKMRKSLKHCIWNVTPSPPRRENDDLEDYGKPEEISEIIDVKKDKEIKQ 536
Query: 654 KTKPIPPS-----SASDYHQNRHRSERPSPDS---SHREVEPPRPTKRKSDHSEREREDR 705
K K S S S+ + R +S + S +S S E E +RKS D
Sbjct: 537 KAKSESESEESKESDSELRKKRRKSYKKSRESDSESESESEVEDRKRRKSRKYSESDSDT 716
Query: 706 DRDYDYHDRHQDRDYXHDRHQHRRHHHRTEASSKKSTDPVT----KSTSRKSSEPDIKSK 761
+ + + R + R R + R R S + TD + RK S+ KSK
Sbjct: 717 NSEEEDRKRRRRRKSSRRRRKSSRKKRRCSDSDESETDDESGYDDSGRKRKRSKRSRKSK 896
Query: 762 SRKSSEPVTK 771
+KSSEPV++
Sbjct: 897 -KKSSEPVSE 923
>TC81466 similar to GP|5821143|dbj|BAA83713.1 RNA binding protein {Homo
sapiens}, partial (3%)
Length = 1109
Score = 43.9 bits (102), Expect = 3e-04
Identities = 48/187 (25%), Positives = 70/187 (36%), Gaps = 10/187 (5%)
Frame = +1
Query: 617 DARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQNRHRSERP 676
D RRKR + + + +GD+ +I P S N+ + P
Sbjct: 1 DERRKRSGPQDIRK------------RQMGDIKAILGGGGDRPRRSREK--SNKREASPP 138
Query: 677 SPDSSHREVEPPRPTKR----KSDHSEREREDRDRDYDYHDRHQDRDYXHDRHQHRRHHH 732
S R P R +R + D SE +REDR R R + D + RRH H
Sbjct: 139 RRRRSRRSESPERSGRRHHRSRRDESESDREDRRRHKGDRRRSRSPD-----DRSRRHRH 303
Query: 733 RTEASSK---KSTDPVTKSTSRKSSEPDIKS---KSRKSSEPVTKSLSLSKTAPTTSAEA 786
R+ SK ST K R + S +SR+ K S+ ++ T
Sbjct: 304 RSPLGSKDLISSTRRTDKHRLRNGDDGASASWSHRSRRDMSKERKGFSIKGSSRRTQKRD 483
Query: 787 AAAAAAD 793
+ AAA+D
Sbjct: 484 SPAAASD 504
>BQ137346 homologue to GP|15027095|em hypothetical protein LT.09 {Leishmania
major}, partial (0%)
Length = 1101
Score = 42.4 bits (98), Expect = 8e-04
Identities = 41/204 (20%), Positives = 79/204 (38%), Gaps = 24/204 (11%)
Frame = +3
Query: 604 PAMSREEFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSA 663
PA + + ++ +R + E E + ++ RD R + + S +T P+
Sbjct: 480 PAQQGNDRKKKQPASRGRAEEENEDKNRHNRARD-ERHDARTHEKDSRTEQT----PTHT 644
Query: 664 SDYHQNRHRSERPSPDSSH------------REVEPPRPTKR------KSDHSEREREDR 705
H + R+ R P+ +H R +PP+PTKR D +R E
Sbjct: 645 RQRHTTKRRTRRHEPEEAHTEETTPTTRTKTRRPQPPKPTKRGRGQAQAPDEKKRATEQN 824
Query: 706 DRDYDYHDRHQDRDYXHDRHQHRRHH-HRTEASSKKSTD-----PVTKSTSRKSSEPDIK 759
+ + + + R + R R H H T ++K+ D T+ +R+ +
Sbjct: 825 ESEREENTRRHTETHDDKRPTAAREHPHNTRETTKRRRDKSAEKQETRERTRRHTTTHTD 1004
Query: 760 SKSRKSSEPVTKSLSLSKTAPTTS 783
+ ++ +P + + AP TS
Sbjct: 1005DQDARTLKPQRTRHAEREDAPKTS 1076
Score = 40.8 bits (94), Expect = 0.002
Identities = 54/252 (21%), Positives = 91/252 (35%), Gaps = 21/252 (8%)
Frame = +1
Query: 614 RKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQNRHRS 673
RK RR+R +K R G++ + ++ K+K S +
Sbjct: 328 RKTTPRRRR----------NKQRTTGKQPKTKAGENARKTKEGQTGEPSGEKPDKGARNR 477
Query: 674 ERPSPDSSHREVEPP------RPTKRKSDHSERERE-----------DRDRDYDYHDRHQ 716
+RP+ ++ + PP R T+ K+D +ERE +++R HDR
Sbjct: 478 DRPNRGTTGKRNNPPAGGAPKRKTRTKTDTTERETRGTTRGRTRRTAEQNRHRHTHDRDT 657
Query: 717 DRDYXHDRHQHRRHHHRTEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTKSLSLS 776
R + R+H + +R+S++ D + E +K +
Sbjct: 658 QRRDEREDTSQRKHTQKKPRQPHAQKRADRNRRNRRSADADKHKHPTRRKEQQSKMRARE 837
Query: 777 KTAPTTSAEAAAAAAADRKQKASV-FSRISFPSEEEAAAAAKKRKLSASSTTEASTAATA 835
K T DR+ +AS ++ S EE A K+ SA + T T T
Sbjct: 838 K--KTRDDTRRRTTTNDRQPRASTRTTQERQRSAEETRAQKNKKHESARADTPRHTQTTK 1011
Query: 836 SV---SAKAPST 844
+ S AP T
Sbjct: 1012TREH*SRNAPGT 1047
>BQ151301 similar to PIR|S34666|S346 glycine-rich protein - common tobacco,
partial (22%)
Length = 530
Score = 42.4 bits (98), Expect = 8e-04
Identities = 37/120 (30%), Positives = 45/120 (36%), Gaps = 6/120 (5%)
Frame = +2
Query: 478 PTEAGKKKKKKKVRMPTNDFQWKPPHDLGAENYMMQMGPPPGYNPYWNGMQPCMDGFMAP 537
PT +K+KKKK P PPH+ GA +M PPP P W+ P P
Sbjct: 77 PTPPPQKQKKKKPHHPPPP----PPHEAGA--VIMPPPPPPPPPPTWHPPPP-------P 217
Query: 538 YAGPMH------MMDYGHGPYDMPFPNGMPHDPFANGMPHDPFGMQGYMMPPIPPPPHRD 591
P H + + H PFP G +G PP PPPPH D
Sbjct: 218 PTPPTHPPPTLFFLFFSH-----PFPGKKGGGGGGEGKKE----RKGNKRPPPPPPPHHD 370
>BQ137267 similar to GP|9800294|gb|A pR79 {rat cytomegalovirus Maastricht},
partial (9%)
Length = 1071
Score = 42.0 bits (97), Expect = 0.001
Identities = 50/248 (20%), Positives = 96/248 (38%), Gaps = 22/248 (8%)
Frame = +1
Query: 683 REVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDRHQHRRHHHRTEASSKKST 742
+++ P PT + + + + + + + R+ H R RR H R + +
Sbjct: 265 KQITPNAPTAHQQADKQLQADTK*TTKTSKNDNTPRERAHPR---RRAHRRRSDENVRGG 435
Query: 743 DPVTKSTSRKSSEPDIKS-KSRKSSEPVTKSLSLSKTAP------TTSAEAAAAAAADRK 795
D + +R P + ++RK + T++ + + T P T E + A+ + +
Sbjct: 436 DTLG---ARSCGPPHADTLRARKLNRRTTQTRASAHTQPRSNQHQTARTERSNASHTNER 606
Query: 796 QKASVFSRISFPSEEEAAAAAKKRKLSASSTTEASTAATASVSAKAPS-----TTHLSNG 850
A SR S +E ++R + T S +TA+ + + T+H ++
Sbjct: 607 TSARALSR----SRDEDVRDTRQRSTRRGAQTRKSPPSTAARATTVRAR*RDRTSHRTHL 774
Query: 851 GRKSKAVMDDYESSDDDEDDERHFKRR----------PSRYEPSPPPPVDDWVEEGRHSR 900
R ++A D +D ER +R P +PP D+ +EGR +
Sbjct: 775 ARDTRA-SDRQRRRTEDATSERE*SQREDIRSRSSHPPRARHAAPPTRDDEMYDEGRREQ 951
Query: 901 GTRDRKHR 908
G D R
Sbjct: 952 GADDESQR 975
Score = 40.8 bits (94), Expect = 0.002
Identities = 57/252 (22%), Positives = 97/252 (37%), Gaps = 13/252 (5%)
Frame = +2
Query: 667 HQNRHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDRHQ 726
+Q ++ P P + R + K++ RERE D D ++ R
Sbjct: 59 NQTQNHDPAPPPPPNPRARRGQERRRDKNNKETRERETGGTDGGRRDPRWNKQDETARAS 238
Query: 727 HRRHHHRTEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTKSLS-LSKTAPTTSAE 785
RR TE SK P + R ++ +K ++ ++ T+ S L++ T E
Sbjct: 239 TRR--DGTE*ESK*RPTPPPPTNRRTNNCKQTPNKQQRQAKTTTRPASALTRGDEHTEDE 412
Query: 786 AAAAAAADRKQKASVFSRISFPSEEEAAAAAKKRKLSASSTTEASTAATASVSAKAP--- 842
A A + + R + A++ + R+ A T + A+++ K P
Sbjct: 413 ATRTYVAATRSARAHAGRPTPTRCARASSIGEPRR-----RERAHTHSHAAINTKQPERN 577
Query: 843 -STTHLSNGGRKSKAVMD-DYESSDDDEDDERHFKRRPSRYEPSPP---PPVDDWVEEGR 897
+T H GR + + + +S+ R +RR +R P PP PP GR
Sbjct: 578 EATHHTLTRGRAREHYREAETRTSETPASAARGEERRHARARPPPPRARPPSAQGSATGR 757
Query: 898 H----SRGTRDR 905
H S TR+R
Sbjct: 758 HIARTSHATRER 793
>TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (91%)
Length = 860
Score = 42.0 bits (97), Expect = 0.001
Identities = 25/68 (36%), Positives = 30/68 (43%)
Frame = +3
Query: 185 GAGRGFGRGMGGRMGGGRGFGLERKTPPQGYVCHRCKVPGHFIQHCPTNGDPNYDVKRVK 244
G G G GRG GG GGG G L+ C+ C PGHF + C G +R +
Sbjct: 273 GGGGGGGRGRGG--GGGGGSDLK---------CYECGEPGHFARECRNRGGGGAGRRRSR 419
Query: 245 QPTGIPRS 252
P RS
Sbjct: 420 SPPRFRRS 443
>TC78386 similar to SP|Q9NZW4|DSPP_HUMAN Dentin sialophosphoprotein
precursor [Contains: Dentin phosphoprotein (Dentin
phosphophoryn) (DPP);, partial (2%)
Length = 726
Score = 41.6 bits (96), Expect = 0.001
Identities = 49/185 (26%), Positives = 82/185 (43%), Gaps = 3/185 (1%)
Frame = +3
Query: 605 AMSREEFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSAS 664
A S + + RRKR + R +RD +KD R+ S S K + K SS S
Sbjct: 69 AASSSSLSSDSSSHRRKRHHRSRRDRD-NKDSLKIRKKSK--SHSHAKRRRKHHRYSSDS 239
Query: 665 DYHQNRHRSERP-SPDSSHREVEPPRPTKR--KSDHSEREREDRDRDYDYHDRHQDRDYX 721
D++ + S+ S SS+ E E +K+ KSD ++ +E ++R
Sbjct: 240 DFYSSSSLSDSSRSESSSNSEHETSHRSKKHKKSDKPKKNKE------------KERSKS 383
Query: 722 HDRHQHRRHHHRTEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTKSLSLSKTAPT 781
H RH+ +H + + + ++S+ PV S ++ D +S S + + L L KT
Sbjct: 384 H-RHKRHKHSLKEKPNGERSSSPVQLSKFLGRNKDDGVRRSAVSGKKIL--LKLDKTKED 554
Query: 782 TSAEA 786
AE+
Sbjct: 555 KEAES 569
Score = 39.7 bits (91), Expect = 0.005
Identities = 37/123 (30%), Positives = 50/123 (40%), Gaps = 6/123 (4%)
Frame = +3
Query: 656 KPIPPSSASDYHQNRHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRH 715
KP+ P+ S H S S SSHR RK H R R DRD R
Sbjct: 36 KPLFPNRFS--HMAASSSSLSSDSSSHR---------RKRHH--RSRRDRDNKDSLKIRK 176
Query: 716 QDRDYXHDRHQHRRHHHRTE------ASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPV 769
+ + + H + RR HHR +SS S ++S+S E +SK K S+
Sbjct: 177 KSKSHSHAKR--RRKHHRYSSDSDFYSSSSLSDSSRSESSSNSEHETSHRSKKHKKSDKP 350
Query: 770 TKS 772
K+
Sbjct: 351 KKN 359
>BQ137243 similar to GP|2624970|gb|A proline-rich protein 9-1 {Mus musculus},
partial (13%)
Length = 1145
Score = 41.6 bits (96), Expect = 0.001
Identities = 39/167 (23%), Positives = 71/167 (42%), Gaps = 5/167 (2%)
Frame = +3
Query: 616 ADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQNRHRSER 675
A R +R ERR+ER+ K R R + D ++ ++ R+ R
Sbjct: 624 AGGRDRRTTERRIEREREKARK-QRNTTITADRGHAHARR-----------DEHTRRASR 767
Query: 676 PSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRH-QDRDYXHDRHQHRRH--HH 732
S R+ E + +R+ D +++RE R + H +H Q +DR + H
Sbjct: 768 RGKQSCRRDREKNQDVQRQRDEHDKQREKRAQ----HRKHTQIECSANDREESEGHEKRR 935
Query: 733 RTEASSKKSTDPVTKSTSRKSSEPDI--KSKSRKSSEPVTKSLSLSK 777
RT S+K++ T S +E D +++R+ P T ++++
Sbjct: 936 RTTESAKEAAQRQTTRPSEHRTESDSTNANEARERQRPATNQTTITR 1076
>BQ137254 similar to GP|22946423|gb| CG31813-PA {Drosophila melanogaster},
partial (12%)
Length = 1220
Score = 41.2 bits (95), Expect = 0.002
Identities = 60/281 (21%), Positives = 99/281 (34%), Gaps = 31/281 (11%)
Frame = +1
Query: 607 SREEFEARKADARRKR-ENERRVERDFSKDRDFGRE----------VSSVGDVSSIKSKT 655
SR++ E R+ ++ R E E+ R S+ R G VG +
Sbjct: 253 SRDQPEDREKGPQKGRGEREQGGRRGPSRTRARGHACQGAEQTEP*ADGVGPAQERRDTH 432
Query: 656 KPIP--PSSASDYHQNRHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHD 713
+P P P+ A Q RH P ++ R + +++ +ERER R R+
Sbjct: 433 RPTPQTPTEAETTKQKRHTERTPPTRATPRRRRERE*REERAETTERERAARRRNTTRAR 612
Query: 714 RHQDRDYXHDRHQHRRHHHRTEASSKKSTDPVTKSTSR---KSSEPDIKSKSRKSSEPVT 770
R R RT +S++++T + R + + D+ R T
Sbjct: 613 GLPQRQQRARARAERSGRQRTHSSTQRTTHARARRARRRDVRDARADVTQGRRNRGRQTT 792
Query: 771 KSL---SLSKTAPTTSAEAAAAAAADRKQKASVFSR----ISFPSEEEAAAAAKK----- 818
S + + TS E A +R QK + +R P A KK
Sbjct: 793 ASARTREQREMSSNTSGERIGTRARERAQKRAAAARARNQTRAPHSRRDDAEGKKAGRPR 972
Query: 819 ---RKLSASSTTEASTAATASVSAKAPSTTHLSNGGRKSKA 856
R + +S + A SV + T N R+++A
Sbjct: 973 HSRRPAAHASDADGQNPAAQSVDGERTRT----NSNRRARA 1083
Score = 30.8 bits (68), Expect = 2.4
Identities = 51/276 (18%), Positives = 101/276 (36%), Gaps = 7/276 (2%)
Frame = +3
Query: 604 PAMSREEFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSA 663
PA +R+ + + + + R ++ R + + +++ + P+ A
Sbjct: 498 PADARDAAQTTRT*VEGRESRDDRARESSAQTEHHTRTRPTTATTARARARGEIGSPAHA 677
Query: 664 SDY--HQNRHRSE----RPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQD 717
+ H R R+ R + +S R+ +P S+ ER +R
Sbjct: 678 QQHAAHHTRARAASTQTRRARRASRRDTRQTQPR*TDDGVSQNERTER------------ 821
Query: 718 RDYXHDRHQHRRHHHRTEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTKSLSLSK 777
D QH R HR +AS++ + S+S ++PD ++ +S + +
Sbjct: 822 -----DEQQHERRAHR-DASTRARAE---ASSSGAGTKPDTRTALAPRRRRRQESGAAAP 974
Query: 778 TAPTTSAEAAAAAAADRKQKASVFSRISFPSEEEAAAAAKKRKLSASSTTEASTAATASV 837
AP A A +R+++ E + A +++R E++ A
Sbjct: 975 LAP-------ACGARERRRR-----------PEPSGAISRRRAHEDEQQQESARTQPAQA 1100
Query: 838 SAKAPSTTHLSNGGRKSKAVMDDYES-SDDDEDDER 872
H +G R +A DD + ++ E DER
Sbjct: 1101QHTLHRDAH-ESGRRAERATSDDRAAHTEASETDER 1205
>TC90482
Length = 454
Score = 41.2 bits (95), Expect = 0.002
Identities = 37/131 (28%), Positives = 48/131 (36%), Gaps = 11/131 (8%)
Frame = +2
Query: 624 NERRVERDF--SKDRDF-------GREVSSVGDVSSIKSKTKPIPPSSAS-DYHQNRHRS 673
+ER VE D KD+D+ GR D ++SK + P DY + H
Sbjct: 11 HERDVEHDRYREKDKDYEVGETDRGRSHDRGADYDHVESKHEKEPHGEIERDYEPDDHHG 190
Query: 674 ERPSPDSSHREVEPPRPTKRKSDHSEREREDRDR-DYDYHDRHQDRDYXHDRHQHRRHHH 732
P HR +P + DH R R YD D D + D H H
Sbjct: 191 RYKEPGHGHRHADP----EHDPDHYNHYEHQRGRGQYDEGDDRGDYNQYPD-HAKMEDDH 355
Query: 733 RTEASSKKSTD 743
TE + KS D
Sbjct: 356 HTERAKSKSRD 388
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.312 0.130 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,790,704
Number of Sequences: 36976
Number of extensions: 497324
Number of successful extensions: 5371
Number of sequences better than 10.0: 280
Number of HSP's better than 10.0 without gapping: 4137
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5018
length of query: 908
length of database: 9,014,727
effective HSP length: 105
effective length of query: 803
effective length of database: 5,132,247
effective search space: 4121194341
effective search space used: 4121194341
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC119408.8


