

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149639.1 + phase: 0
(164 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
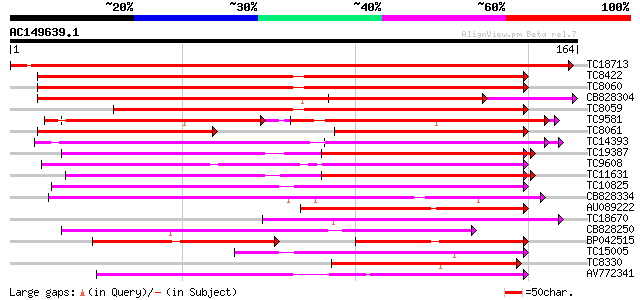
Score E
Sequences producing significant alignments: (bits) Value
TC18713 similar to GB|AAL90989.1|19699246|AY090328 AT3g03000/F13... 284 5e-78
TC8422 UP|AAQ63461 (AAQ63461) Calmodulin 4 (Fragment), complete 139 2e-34
TC8060 UP|O49184 (O49184) Calmodulin (ESTS AU081349) (Auxin-regu... 137 9e-34
CB828304 136 2e-33
TC8059 GB|AAB68399.1|1773321|HAU79736 calmodulin {Helianthus ann... 115 4e-27
TC9581 UP|Q9SCA1 (Q9SCA1) Calcium-binding protein, complete 103 2e-23
TC8061 UP|AAA34013 (AAA34013) Calmodulin, complete 69 2e-22
TC14393 similar to UP|Q93YA8 (Q93YA8) Calcium binding protein, p... 96 3e-21
TC19387 weakly similar to UP|Q9FYK2 (Q9FYK2) F21J9.28, partial (... 95 7e-21
TC9608 similar to UP|Q94AZ4 (Q94AZ4) At1g12310/F5O11_2, partial ... 94 2e-20
TC11631 weakly similar to UP|TCH2_ARATH (P25070) Calmodulin-rela... 86 3e-18
TC10825 similar to GB|AAP68339.1|31711966|BT008900 At1g74740 {Ar... 84 9e-18
CB828334 61 8e-11
AU089222 56 4e-09
TC18670 similar to UP|Q93YA8 (Q93YA8) Calcium binding protein, p... 55 8e-09
CB828250 54 2e-08
BP042515 44 1e-05
TC15005 homologue to UP|Q9AR92 (Q9AR92) Protein kinase , partia... 43 3e-05
TC8330 weakly similar to GB|AAM61257.1|21536925|AY084696 calmodu... 42 4e-05
AV772341 38 8e-04
>TC18713 similar to GB|AAL90989.1|19699246|AY090328 AT3g03000/F13E7_5
{Arabidopsis thaliana;}, partial (88%)
Length = 868
Score = 284 bits (727), Expect = 5e-78
Identities = 144/163 (88%), Positives = 157/163 (95%)
Frame = +1
Query: 1 MNKKQHQVKLDDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQR 60
M+KKQ V LD+EQI+ELREIFRSFDRNNDG+LTQLEL+SLLRSLGLKPSAEQ+EGFIQ+
Sbjct: 169 MSKKQ-AVNLDEEQIAELREIFRSFDRNNDGSLTQLELSSLLRSLGLKPSAEQVEGFIQK 345
Query: 61 ADTNNNGLIEFSEFVALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAK 120
ADTN+NGLIEFSEFVA+VAPELLPA+SPYTEEQLRQLFR+FDRDGNGFITAAELAHSMAK
Sbjct: 346 ADTNSNGLIEFSEFVAVVAPELLPARSPYTEEQLRQLFRVFDRDGNGFITAAELAHSMAK 525
Query: 121 LGHALTAEELTGMIKEADMDGDGMISFQEFGQAITSAAFDNSW 163
LGHAL+AEELTGMIKEAD DGDGMISFQEF AI+SAAFDNSW
Sbjct: 526 LGHALSAEELTGMIKEADADGDGMISFQEFAHAISSAAFDNSW 654
>TC8422 UP|AAQ63461 (AAQ63461) Calmodulin 4 (Fragment), complete
Length = 832
Score = 139 bits (350), Expect = 2e-34
Identities = 68/142 (47%), Positives = 100/142 (69%)
Frame = +1
Query: 9 KLDDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGL 68
+L D+QISE +E F FD++ DG++T EL +++RSLG P+ +L+ I D + NG
Sbjct: 145 QLTDDQISEFKEAFSLFDKDGDGSITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGT 324
Query: 69 IEFSEFVALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAE 128
I+F EF+ L+A ++ K +EE+L++ FR+FD+D NGFI+AAEL H M LG LT E
Sbjct: 325 IDFPEFLNLMARKM---KDTDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDE 495
Query: 129 ELTGMIKEADMDGDGMISFQEF 150
E+ MI+EAD+DGDG I+++EF
Sbjct: 496 EVDEMIREADVDGDGQINYEEF 561
>TC8060 UP|O49184 (O49184) Calmodulin (ESTS AU081349) (Auxin-regulated
calmodulin) (Calmodulin TACAM1-1) (Calmodulin TACAM1-2)
(Calmodulin TACAM1-3) (Calmodulin TACAM3-1) (Calmodulin
TACAM3-2) (Calmodulin TACAM3-3) (Calmodulin TACAM4-1)
(CaM protein), complete
Length = 615
Score = 137 bits (345), Expect = 9e-34
Identities = 67/142 (47%), Positives = 99/142 (69%)
Frame = +3
Query: 9 KLDDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGL 68
+L D+QI+E +E F FD++ DG +T EL +++RSLG P+ +L+ I D + NG
Sbjct: 78 QLTDDQIAEFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGT 257
Query: 69 IEFSEFVALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAE 128
I+F EF+ L+A ++ K +EE+L++ FR+FD+D NGFI+AAEL H M LG LT E
Sbjct: 258 IDFPEFLNLMARKM---KDTDSEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDE 428
Query: 129 ELTGMIKEADMDGDGMISFQEF 150
E+ MI+EAD+DGDG I+++EF
Sbjct: 429 EVDEMIREADVDGDGQINYEEF 494
>CB828304
Length = 489
Score = 136 bits (342), Expect = 2e-33
Identities = 71/131 (54%), Positives = 94/131 (71%), Gaps = 1/131 (0%)
Frame = +1
Query: 9 KLDDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGL 68
KL +Q+++LREIF FD ++DG+LT LEL +LLRSLGLKPS +QL + D+N NG
Sbjct: 94 KLQVQQLNQLREIFARFDMDSDGSLTMLELAALLRSLGLKPSGDQLHDLLSNMDSNGNGS 273
Query: 69 IEFSEFVALVAPELL-PAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTA 127
+EF E V + P+L A+ +EQL +F+ FDRD NGFI+AAELA +MAK+G LT
Sbjct: 274 VEFDELVRTILPDLKNNAEVLLNQEQLLDVFKCFDRDSNGFISAAELAGAMAKMGQPLTY 453
Query: 128 EELTGMIKEAD 138
+ELT MI+EAD
Sbjct: 454 KELTEMIREAD 486
Score = 45.4 bits (106), Expect = 5e-06
Identities = 23/72 (31%), Positives = 39/72 (53%)
Frame = +1
Query: 93 QLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAEELTGMIKEADMDGDGMISFQEFGQ 152
QLR++F FD D +G +T ELA + LG + ++L ++ D +G+G + F E +
Sbjct: 118 QLREIFARFDMDSDGSLTMLELAALLRSLGLKPSGDQLHDLLSNMDSNGNGSVEFDELVR 297
Query: 153 AITSAAFDNSWV 164
I +N+ V
Sbjct: 298 TILPDLKNNAEV 333
>TC8059 GB|AAB68399.1|1773321|HAU79736 calmodulin {Helianthus annuus;} ,
partial (83%)
Length = 836
Score = 115 bits (288), Expect = 4e-27
Identities = 57/120 (47%), Positives = 83/120 (68%)
Frame = +2
Query: 31 GTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLIEFSEFVALVAPELLPAKSPYT 90
G +T EL +++RSLG P+ +L+ I D + NG I+F EF+ L+A ++ K +
Sbjct: 164 GCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLNLMARKM---KDTDS 334
Query: 91 EEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAEELTGMIKEADMDGDGMISFQEF 150
EE+L++ FR+FD+D NGFI+AAEL H M LG LT EE+ MI+EAD+DGDG I+++EF
Sbjct: 335 EEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIREADVDGDGQINYEEF 514
>TC9581 UP|Q9SCA1 (Q9SCA1) Calcium-binding protein, complete
Length = 1049
Score = 103 bits (256), Expect = 2e-23
Identities = 55/146 (37%), Positives = 86/146 (58%), Gaps = 2/146 (1%)
Frame = +3
Query: 16 SELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLIEFSEFV 75
+EL+ +F+ FDRN DG +T+ ELN L +LG+ ++L I+R D N +G ++ EF
Sbjct: 345 TELKRVFQMFDRNGDGRITKKELNDSLENLGIFIPDKELTQMIERIDVNGDGCVDIDEFG 524
Query: 76 ALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLG--HALTAEELTGM 133
L ++ + EE +R+ F +FD++G+GFIT EL +A LG T E+ M
Sbjct: 525 ELY-QSIMDERD--EEEDMREAFNVFDQNGDGFITVEELRTVLASLGIKQGRTVEDCKKM 695
Query: 134 IKEADMDGDGMISFQEFGQAITSAAF 159
I + D+DGDGM+ ++EF Q + F
Sbjct: 696 IMKVDVDGDGMVDYKEFKQMMKGGGF 773
Score = 60.5 bits (145), Expect = 1e-10
Identities = 31/75 (41%), Positives = 47/75 (62%)
Frame = +3
Query: 82 LLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAEELTGMIKEADMDG 141
L+P P +L+++F+MFDR+G+G IT EL S+ LG + +ELT MI+ D++G
Sbjct: 324 LIPKMDP---TELKRVFQMFDRNGDGRITKKELNDSLENLGIFIPDKELTQMIERIDVNG 494
Query: 142 DGMISFQEFGQAITS 156
DG + EFG+ S
Sbjct: 495 DGCVDIDEFGELYQS 539
Score = 49.3 bits (116), Expect = 3e-07
Identities = 24/66 (36%), Positives = 42/66 (63%), Gaps = 2/66 (3%)
Frame = +3
Query: 11 DDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKP--SAEQLEGFIQRADTNNNGL 68
D+E+ ++RE F FD+N DG +T EL ++L SLG+K + E + I + D + +G+
Sbjct: 555 DEEE--DMREAFNVFDQNGDGFITVEELRTVLASLGIKQGRTVEDCKKMIMKVDVDGDGM 728
Query: 69 IEFSEF 74
+++ EF
Sbjct: 729 VDYKEF 746
>TC8061 UP|AAA34013 (AAA34013) Calmodulin, complete
Length = 568
Score = 68.9 bits (167), Expect(2) = 2e-22
Identities = 32/56 (57%), Positives = 42/56 (74%)
Frame = +2
Query: 95 RQLFRMFDRDGNGFITAAELAHSMAKLGHALTAEELTGMIKEADMDGDGMISFQEF 150
R+L + D+D NGFI+AAEL H M LG LT EE+ MI+EAD+DGDG I+++EF
Sbjct: 305 RKLSALLDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIREADVDGDGQINYEEF 472
Score = 51.6 bits (122), Expect(2) = 2e-22
Identities = 24/52 (46%), Positives = 35/52 (67%)
Frame = +3
Query: 9 KLDDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQR 60
+L DEQISE +E F FD++ DG +T EL +++RSLG P+ +L+ QR
Sbjct: 54 QLTDEQISEFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAELQAHDQR 209
Score = 36.6 bits (83), Expect = 0.002
Identities = 20/50 (40%), Positives = 27/50 (54%), Gaps = 3/50 (6%)
Frame = +3
Query: 84 PAKSPYTEEQL---RQLFRMFDRDGNGFITAAELAHSMAKLGHALTAEEL 130
P T+EQ+ ++ F +FD+DG+G IT EL M LG T EL
Sbjct: 42 PMADQLTDEQISEFKEAFSLFDKDGDGCITTKELGTVMRSLGQNPTEAEL 191
>TC14393 similar to UP|Q93YA8 (Q93YA8) Calcium binding protein, partial
(85%)
Length = 786
Score = 95.9 bits (237), Expect = 3e-21
Identities = 55/149 (36%), Positives = 85/149 (56%)
Frame = +3
Query: 8 VKLDDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNG 67
V LDD + EL+ +F FD N DG ++ EL+++LRSLG E+L+ + D +++G
Sbjct: 129 VYLDD--MEELKRVFTRFDTNGDGKISVNELDNVLRSLGSGVPPEELKRVMVDLDGDHDG 302
Query: 68 LIEFSEFVALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTA 127
I SEF A + + +L F ++D+D NG I+AAEL + +LG +
Sbjct: 303 FINLSEFAAFCRSDTADGGA----SELHDAFELYDQDKNGLISAAELCKVLNRLGMNCSE 470
Query: 128 EELTGMIKEADMDGDGMISFQEFGQAITS 156
EE MIK D DGDG ++F+EF + +T+
Sbjct: 471 EECHKMIKSVDSDGDGNVNFEEFKKMMTN 557
Score = 45.8 bits (107), Expect = 4e-06
Identities = 24/69 (34%), Positives = 38/69 (54%)
Frame = +3
Query: 92 EQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAEELTGMIKEADMDGDGMISFQEFG 151
E+L+++F FD +G+G I+ EL + + LG + EEL ++ + D D DG I+ EF
Sbjct: 147 EELKRVFTRFDTNGDGKISVNELDNVLRSLGSGVPPEELKRVMVDLDGDHDGFINLSEFA 326
Query: 152 QAITSAAFD 160
S D
Sbjct: 327 AFCRSDTAD 353
>TC19387 weakly similar to UP|Q9FYK2 (Q9FYK2) F21J9.28, partial (45%)
Length = 631
Score = 94.7 bits (234), Expect = 7e-21
Identities = 51/135 (37%), Positives = 76/135 (55%)
Frame = +1
Query: 16 SELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLIEFSEFV 75
+E+REIF+ FDRN DG ++ EL L+ +LG K +AE++ + D N +G I+ EF
Sbjct: 43 NEVREIFKKFDRNGDGKISCSELKDLMAALGSKTTAEEVRRMMAELDRNGDGHIDLKEF- 219
Query: 76 ALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAEELTGMIK 135
E +++R+ F M+D D NG I+A EL M KLG + + MI
Sbjct: 220 ----GEFHRGGGGGNSKEIREAFEMYDLDKNGLISAKELHSVMKKLGEKCSLGDCRKMIG 387
Query: 136 EADMDGDGMISFQEF 150
D+DGDG ++F+EF
Sbjct: 388 NVDVDGDGNVNFEEF 432
Score = 61.2 bits (147), Expect = 8e-11
Identities = 28/62 (45%), Positives = 43/62 (69%)
Frame = +1
Query: 91 EEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAEELTGMIKEADMDGDGMISFQEF 150
+ ++R++F+ FDR+G+G I+ +EL MA LG TAEE+ M+ E D +GDG I +EF
Sbjct: 40 DNEVREIFKKFDRNGDGKISCSELKDLMAALGSKTTAEEVRRMMAELDRNGDGHIDLKEF 219
Query: 151 GQ 152
G+
Sbjct: 220 GE 225
>TC9608 similar to UP|Q94AZ4 (Q94AZ4) At1g12310/F5O11_2, partial (97%)
Length = 817
Score = 93.6 bits (231), Expect = 2e-20
Identities = 49/141 (34%), Positives = 78/141 (54%)
Frame = +3
Query: 10 LDDEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLI 69
L D+Q+S ++E F FD + DG + EL L+RSLG P+ QL+ + A+ N
Sbjct: 117 LSDDQVSSMKEAFTLFDTDGDGKIAPSELGILMRSLGGNPTQAQLKSIV--AEENLTSPF 290
Query: 70 EFSEFVALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAEE 129
+F F+ L+A + P P+ + QLR F++ D+D GF++ EL H + +G L E
Sbjct: 291 DFPRFLDLMAKHMKP--EPF-DRQLRDAFKVIDKDSTGFVSVTELRHILTSIGEKLEPAE 461
Query: 130 LTGMIKEADMDGDGMISFQEF 150
I+E D+ DG I +++F
Sbjct: 462 FDEWIREVDVGSDGKIRYEDF 524
>TC11631 weakly similar to UP|TCH2_ARATH (P25070) Calmodulin-related protein
2, touch-induced, partial (68%)
Length = 530
Score = 85.9 bits (211), Expect = 3e-18
Identities = 48/134 (35%), Positives = 73/134 (53%)
Frame = +3
Query: 17 ELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLIEFSEFVA 76
E+R+IF FD+N DG ++ EL +L +LG K S+E+++ + D N +G I+ EF
Sbjct: 99 EVRKIFNKFDKNGDGKISCTELKEMLCALGTKTSSEEVKRMMAEIDQNGDGYIDLKEF-- 272
Query: 77 LVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAEELTGMIKE 136
A L ++LR ++D D NG I+A EL + KLG + + MI
Sbjct: 273 --AEFHLHDAGEDDSKELRDACDLYDLDKNGLISANELHSVLKKLGEKCSLGDCRKMISN 446
Query: 137 ADMDGDGMISFQEF 150
D DGDG ++F+EF
Sbjct: 447 VDADGDGNVNFEEF 488
Score = 52.8 bits (125), Expect = 3e-08
Identities = 22/62 (35%), Positives = 40/62 (64%)
Frame = +3
Query: 91 EEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAEELTGMIKEADMDGDGMISFQEF 150
++++R++F FD++G+G I+ EL + LG ++EE+ M+ E D +GDG I +EF
Sbjct: 93 DDEVRKIFNKFDKNGDGKISCTELKEMLCALGTKTSSEEVKRMMAEIDQNGDGYIDLKEF 272
Query: 151 GQ 152
+
Sbjct: 273 AE 278
>TC10825 similar to GB|AAP68339.1|31711966|BT008900 At1g74740 {Arabidopsis
thaliana;}, partial (32%)
Length = 1091
Score = 84.3 bits (207), Expect = 9e-18
Identities = 40/138 (28%), Positives = 82/138 (58%)
Frame = +1
Query: 13 EQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLIEFS 72
E++ ++++F D + DG ++ EL + LR +G + + ++++ ++ AD + NG++++
Sbjct: 4 EEVEIIKDMFTLMDTDKDGRVSYEELKAGLRKVGSQLADQEIKMLMEVADVDGNGVLDYG 183
Query: 73 EFVALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAEELTG 132
EFVA+ + + +E + F+ FD+DG+G+I + EL ++A A+ L
Sbjct: 184 EFVAVT----IHLQKMENDEHFHKAFKFFDKDGSGYIESGELQEALADESGVTDADVLND 351
Query: 133 MIKEADMDGDGMISFQEF 150
+++E D D DG IS++EF
Sbjct: 352 IMREVDTDKDGRISYEEF 405
>CB828334
Length = 541
Score = 61.2 bits (147), Expect = 8e-11
Identities = 43/164 (26%), Positives = 78/164 (47%), Gaps = 20/164 (12%)
Frame = +2
Query: 12 DEQISELREIFRSFDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLIEF 71
DE + + IF+ FD +++G + Q EL L + + E++ + D N + +EF
Sbjct: 35 DESFGKCKAIFKQFDEDSNGAIDQEELKKCFSKLEISFTEEEVNDLFEACDINEDMGMEF 214
Query: 72 SEFVALVA--------PELLPAKS-------PYTEEQLRQLFRMFDRDGNGFITAAELAH 116
SEF+ L+ + AKS T E L F D++ +G+++ +E+
Sbjct: 215 SEFIVLLCLVHLLKDDRAAVQAKSRIGMPDLETTFETLVDTFVFLDKNKDGYVSKSEMVQ 394
Query: 117 SMAKLGHALTAEELTGMI-----KEADMDGDGMISFQEFGQAIT 155
+ + ++ E +G I +E D D +GM++F+EF A T
Sbjct: 395 A---INETVSGERSSGRIAMKRFEEMDWDKNGMVNFKEFLFAFT 517
>AU089222
Length = 345
Score = 55.8 bits (133), Expect = 4e-09
Identities = 27/66 (40%), Positives = 44/66 (65%)
Frame = +3
Query: 85 AKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAEELTGMIKEADMDGDGM 144
A P ++F+ FD +G+G I+++EL ++ LG ++TAEE+ M+ E D+DGDG
Sbjct: 51 ADDPQDVADRERIFKRFDANGDGQISSSELGEALKTLG-SVTAEEVQRMMDEIDIDGDGF 227
Query: 145 ISFQEF 150
IS++EF
Sbjct: 228 ISYEEF 245
Score = 31.2 bits (69), Expect = 0.093
Identities = 22/74 (29%), Positives = 33/74 (43%)
Frame = +3
Query: 55 EGFIQRADTNNNGLIEFSEFVALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAEL 114
E +R D N +G I SE E L T E+++++ D DG+GFI+ E
Sbjct: 81 ERIFKRFDANGDGQISSSEL-----GEALKTLGSVTAEEVQRMMDEIDIDGDGFISYEEF 245
Query: 115 AHSMAKLGHALTAE 128
S A+ L +
Sbjct: 246 T-SFARANRGLVKD 284
>TC18670 similar to UP|Q93YA8 (Q93YA8) Calcium binding protein, partial
(41%)
Length = 329
Score = 54.7 bits (130), Expect = 8e-09
Identities = 33/89 (37%), Positives = 46/89 (51%), Gaps = 2/89 (2%)
Frame = +3
Query: 74 FVALVAPELLPAKSPYTEE--QLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAEELT 131
F A PEL P SPY + +L ++F FD + +G I+ EL + LG + EEL
Sbjct: 51 FPAKTDPELPPTASPYLQNTAELERVFNRFDANSDGKISVDELDSVLRTLGSGVPPEELQ 230
Query: 132 GMIKEADMDGDGMISFQEFGQAITSAAFD 160
++++ D D DG IS EF S A D
Sbjct: 231 QVMEDLDTDRDGFISLPEFAAFCRSDAPD 317
>CB828250
Length = 485
Score = 53.5 bits (127), Expect = 2e-08
Identities = 33/122 (27%), Positives = 61/122 (49%), Gaps = 2/122 (1%)
Frame = +1
Query: 16 SELREIFRSFDRNNDGTLTQLELNSLLRSL--GLKPSAEQLEGFIQRADTNNNGLIEFSE 73
S+ R F D + DG +++ +L + + G+ + + + ADT+ +G +E+ E
Sbjct: 112 SDFRPAFDVLDADCDGKISRDDLRAFYAGIYGGVASGDDVIGAMMTVADTDKDGFVEYEE 291
Query: 74 FVALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAEELTGM 133
F +V P EE +F++ D+DG+G ++ +L MA G + T E++ M
Sbjct: 292 FERVVTENNKPLGCGAMEE----VFKVMDKDGDGKLSHHDLKSYMASAGFSATDEDINAM 459
Query: 134 IK 135
IK
Sbjct: 460 IK 465
Score = 30.8 bits (68), Expect = 0.12
Identities = 16/69 (23%), Positives = 30/69 (43%)
Frame = +1
Query: 91 EEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAEELTGMIKEADMDGDGMISFQEF 150
++ + + + D D +GF+ E + + L + + K D DGDG +S +
Sbjct: 223 DDVIGAMMTVADTDKDGFVEYEEFERVVTENNKPLGCGAMEEVFKVMDKDGDGKLSHHDL 402
Query: 151 GQAITSAAF 159
+ SA F
Sbjct: 403 KSYMASAGF 429
>BP042515
Length = 539
Score = 44.3 bits (103), Expect = 1e-05
Identities = 23/50 (46%), Positives = 30/50 (60%)
Frame = -1
Query: 101 FDRDGNGFITAAELAHSMAKLGHALTAEELTGMIKEADMDGDGMISFQEF 150
FD+DG+G+IT EL + + G L L MI+EAD D DG I + EF
Sbjct: 539 FDKDGSGYITQDELQQACEEFG--LGDVRLEEMIREADQDNDGRIDYNEF 396
Score = 40.4 bits (93), Expect = 2e-04
Identities = 21/54 (38%), Positives = 33/54 (60%)
Frame = -1
Query: 25 FDRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLIEFSEFVALV 78
FD++ G +TQ EL GL +LE I+ AD +N+G I+++EFVA++
Sbjct: 539 FDKDGSGYITQDELQQACEEFGL--GDVRLEEMIREADQDNDGRIDYNEFVAMM 384
>TC15005 homologue to UP|Q9AR92 (Q9AR92) Protein kinase , partial (19%)
Length = 657
Score = 42.7 bits (99), Expect = 3e-05
Identities = 29/91 (31%), Positives = 44/91 (47%), Gaps = 6/91 (6%)
Frame = +2
Query: 66 NGLIEFSEFVALVAPELLPAKSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHAL 125
+G I++ EFV + ++ L F FD+D +GFIT EL +M + G
Sbjct: 14 DGTIDYIEFVTAT----MHRHKLERDDHLN*AFPYFDKDSSGFITRDELEIAMKEYGMGD 181
Query: 126 TA------EELTGMIKEADMDGDGMISFQEF 150
A E+ +I E D D DG I+++EF
Sbjct: 182 DATIKEIISEVDTIISEVDTDHDGRINYEEF 274
>TC8330 weakly similar to GB|AAM61257.1|21536925|AY084696 calmodulin-like
protein {Arabidopsis thaliana;} , partial (72%)
Length = 1174
Score = 42.4 bits (98), Expect = 4e-05
Identities = 22/56 (39%), Positives = 35/56 (62%), Gaps = 1/56 (1%)
Frame = +3
Query: 94 LRQLFRMFDRDGNGFITAAELAHSMAKLGH-ALTAEELTGMIKEADMDGDGMISFQ 148
+R FR+ DRD +G ++ A+L + +LG L+ EE+ M++E D DG G IS +
Sbjct: 294 VRWAFRLIDRDNDGVVSRADLVAVLTRLGALQLSDEEVEVMLREVDGDGRGSISVE 461
>AV772341
Length = 513
Score = 38.1 bits (87), Expect = 8e-04
Identities = 28/125 (22%), Positives = 54/125 (42%)
Frame = +3
Query: 26 DRNNDGTLTQLELNSLLRSLGLKPSAEQLEGFIQRADTNNNGLIEFSEFVALVAPELLPA 85
D G++T +L + L L+ ++ I+ D + NG + F EFVAL L
Sbjct: 42 DSEKTGSITAPQLKTALAVGNLEFPLSVVQQMIRMCDFDRNGTMSFQEFVALNTFLL--- 212
Query: 86 KSPYTEEQLRQLFRMFDRDGNGFITAAELAHSMAKLGHALTAEELTGMIKEADMDGDGMI 145
+++ F +R G GF+ ++ ++ K+G L + + + D +G +
Sbjct: 213 -------KVQHAFSDLER-GRGFLVPDDVFEALVKIGFMLDSPAFYSVCESFDQSKNGRL 368
Query: 146 SFQEF 150
+F
Sbjct: 369 RLDDF 383
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.133 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,810,657
Number of Sequences: 28460
Number of extensions: 15866
Number of successful extensions: 131
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 96
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 111
length of query: 164
length of database: 4,897,600
effective HSP length: 83
effective length of query: 81
effective length of database: 2,535,420
effective search space: 205369020
effective search space used: 205369020
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 52 (24.6 bits)
Medicago: description of AC149639.1


