

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149580.1 + phase: 0 /pseudo
(260 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
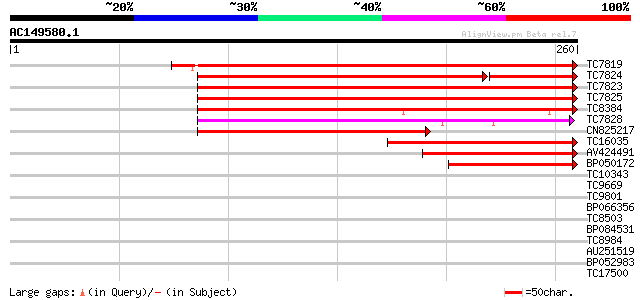
Score E
Sequences producing significant alignments: (bits) Value
TC7819 similar to UP|Q43817 (Q43817) Lipoxygenase , partial (97%) 279 4e-76
TC7824 similar to UP|O04919 (O04919) Lipoxygenase , partial (58%) 213 2e-68
TC7823 similar to UP|O24470 (O24470) Lipoxygenase , partial (96%) 251 9e-68
TC7825 similar to UP|Q9M3Z5 (Q9M3Z5) Lipoxygenase (Fragment) , ... 248 7e-67
TC8384 similar to UP|P93698 (P93698) CPRD46 protein , partial (48%) 197 2e-51
TC7828 similar to UP|O24470 (O24470) Lipoxygenase , partial (48%) 171 1e-43
CN825217 165 8e-42
TC16035 similar to UP|O04919 (O04919) Lipoxygenase , partial (32%) 129 5e-31
AV424491 98 2e-21
BP050172 91 2e-19
TC10343 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (... 35 0.017
TC9669 similar to UP|Q9LQA7 (Q9LQA7) F4N2.10, partial (16%) 34 0.022
TC9801 homologue to UP|Q8LAC6 (Q8LAC6) Glycine rich protein-like... 33 0.049
BP066356 33 0.064
TC8503 similar to GB|AAD30253.1|4835787|F25C20 ESTs gb|R65381 an... 32 0.084
BP084531 32 0.11
TC8984 similar to UP|Q8LBA3 (Q8LBA3) Phosphoserine aminotransfer... 32 0.11
AU251519 32 0.14
BP052983 32 0.14
TC17500 UP|Q9ZPL9 (Q9ZPL9) Nodule-enhanced protein phosphatase t... 31 0.19
>TC7819 similar to UP|Q43817 (Q43817) Lipoxygenase , partial (97%)
Length = 2842
Score = 279 bits (713), Expect = 4e-76
Identities = 133/192 (69%), Positives = 153/192 (79%), Gaps = 6/192 (3%)
Frame = +2
Query: 75 PNSTAYHA------GTFNWLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSV 128
P ST Y GT WLLAKAY +VNDSC HQLVSHWL THAVVEPF+IATNRHLSV
Sbjct: 1538 PESTVYTPAGQGADGTI-WLLAKAYVIVNDSCYHQLVSHWLNTHAVVEPFIIATNRHLSV 1714
Query: 129 LHPIHKLLVPHYRGTMTINARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEG 188
+HPI+KLL PHYR TM INA +RN+L+NA GI+ESTFL+ Y +EMSAV YKDW F E+
Sbjct: 1715 VHPIYKLLFPHYRDTMNINALARNVLVNAEGIIESTFLWGNYALEMSAVAYKDWNFTEQA 1894
Query: 189 LPTDLKNRGMAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVK 248
LP DL RG+AV+D S+PHG+RLLIEDYPYA+DGLEIW AIKSWV EYVN YY+SD ++
Sbjct: 1895 LPADLLKRGVAVEDPSAPHGIRLLIEDYPYAADGLEIWDAIKSWVGEYVNLYYKSDAEIV 2074
Query: 249 DDEELKAFWKEL 260
D EL+AFW EL
Sbjct: 2075 QDAELQAFWTEL 2110
>TC7824 similar to UP|O04919 (O04919) Lipoxygenase , partial (58%)
Length = 1675
Score = 213 bits (542), Expect(2) = 2e-68
Identities = 99/133 (74%), Positives = 114/133 (85%)
Frame = +2
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY +VNDSC HQLVSHWL THAVVEPFVIATNR LSV+HPI+KLL PHYR TM I
Sbjct: 422 WLLAKAYVIVNDSCFHQLVSHWLNTHAVVEPFVIATNRQLSVIHPIYKLLFPHYRDTMNI 601
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R+ L+NA GI+E TFL+ KY MEMS+VVYKDWVF ++ LP DL RG+AV+D S+P
Sbjct: 602 NALARSALVNAEGIIEKTFLWGKYAMEMSSVVYKDWVFTDQALPADLIKRGIAVEDFSAP 781
Query: 207 HGLRLLIEDYPYA 219
HGLRL+IEDYPYA
Sbjct: 782 HGLRLVIEDYPYA 820
Score = 62.4 bits (150), Expect(2) = 2e-68
Identities = 26/40 (65%), Positives = 32/40 (80%)
Frame = +3
Query: 221 DGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
DGL+IWAAIK+WV EYV YY SD ++ D EL+A+WKEL
Sbjct: 825 DGLDIWAAIKTWVQEYVYLYYPSDDAIQQDSELQAWWKEL 944
>TC7823 similar to UP|O24470 (O24470) Lipoxygenase , partial (96%)
Length = 2909
Score = 251 bits (641), Expect = 9e-68
Identities = 116/174 (66%), Positives = 138/174 (78%)
Frame = +1
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY +VNDSC HQL+SHWL THAV+EPFVIATNRHLSVLHPI+KLL PHYR T+ I
Sbjct: 1552 WLLAKAYVIVNDSCYHQLMSHWLNTHAVMEPFVIATNRHLSVLHPIYKLLHPHYRDTINI 1731
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N +R LINAGGI+E +F +EMS+ VYKDWVF ++ LP DL RGMAV+D S+P
Sbjct: 1732 NGLARESLINAGGIIEQSFCPGPNSIEMSSAVYKDWVFTDQALPADLIKRGMAVEDPSAP 1911
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RL++EDYPYA DGLEIW AI WV EYV+ YY +D VK D EL+ +WKE+
Sbjct: 1912 HGVRLVVEDYPYAVDGLEIWDAITKWVQEYVSLYYPTDDAVKKDTELQTWWKEV 2073
>TC7825 similar to UP|Q9M3Z5 (Q9M3Z5) Lipoxygenase (Fragment) , partial
(93%)
Length = 1642
Score = 248 bits (633), Expect = 7e-67
Identities = 115/174 (66%), Positives = 139/174 (79%)
Frame = +1
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKA+ +VNDS HQL+SHWL THA +EPFVIATNRHLSVLHPI KLL PHYR T+ I
Sbjct: 430 WLLAKAHVIVNDSGYHQLISHWLNTHASIEPFVIATNRHLSVLHPIFKLLHPHYRDTINI 609
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N +R LINA GI+E +FL +Y ME+S+VVY+DWVF ++ LP DL RGMAV+D S+P
Sbjct: 610 NGLARQALINADGIIEQSFLPGQYSMEISSVVYRDWVFPDQALPADLIKRGMAVEDPSAP 789
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HGLRL +EDYPYA DGLEIW AIK WV EYV+ YY +D V+ D EL+++WKE+
Sbjct: 790 HGLRLAVEDYPYAVDGLEIWDAIKKWVQEYVSLYYPTDDAVQKDTELQSWWKEV 951
>TC8384 similar to UP|P93698 (P93698) CPRD46 protein , partial (48%)
Length = 1660
Score = 197 bits (500), Expect = 2e-51
Identities = 93/181 (51%), Positives = 129/181 (70%), Gaps = 7/181 (3%)
Frame = +2
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAKA+ + +DS HQLVSHWL+TH EP++IATNR LS +HPI++LL PH+R TM I
Sbjct: 221 WRLAKAHVLAHDSGYHQLVSHWLRTHCATEPYIIATNRQLSAMHPIYRLLHPHFRYTMEI 400
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVY-KDWVFAEEGLPTDLKNRGMAVDDDSS 205
NA +R LINA G++E+ F + +S+V Y + W F + LP DL +RG+A +D ++
Sbjct: 401 NALAREALINANGVIENNFTPGNLSILLSSVAYDQHWQFNLQALPADLIHRGLAEEDPNA 580
Query: 206 PHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKD------VKDDEELKAFWKE 259
PHGL+L IEDYPYA+DGL +W AIKSWV +YVN YY + ++ D+EL+A+W+E
Sbjct: 581 PHGLKLSIEDYPYANDGLILWDAIKSWVTDYVNHYYSEPEANTGPSLIESDQELQAWWEE 760
Query: 260 L 260
+
Sbjct: 761 I 763
>TC7828 similar to UP|O24470 (O24470) Lipoxygenase , partial (48%)
Length = 1371
Score = 171 bits (433), Expect = 1e-43
Identities = 92/197 (46%), Positives = 119/197 (59%), Gaps = 24/197 (12%)
Frame = +2
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKA+ +VNDS HQL++HWL THA +EPF IATNRHLS LHPI+KLL PHYR T+ I
Sbjct: 680 WLLAKAHVIVNDSGYHQLITHWLNTHATIEPFAIATNRHLSALHPINKLLYPHYRDTINI 859
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRG--------- 197
NA +R LINA GI+E +FL +Y +E+S+VVYK+WVF ++ LP DL R
Sbjct: 860 NALARASLINAEGIIEQSFLPGEYSLEISSVVYKNWVFPDQALPADLIKR*HF*I*ITIL 1039
Query: 198 ------MAVDDDSSPHGLRLLIEDYPYAS---------DGLEIWAAIKSWVDEYVNFYYE 242
+ DS LR+ + +A L + WV +YV+ YY
Sbjct: 1040SFLF*LTNILLDSEEWQLRIPLPPMVFALC*RTTLMLLTDLRYGMLLSKWVQDYVSLYYP 1219
Query: 243 SDKDVKDDEELKAFWKE 259
SD V+ D EL+A+WKE
Sbjct: 1220SDDAVQKDTELQAWWKE 1270
>CN825217
Length = 685
Score = 165 bits (417), Expect = 8e-42
Identities = 74/107 (69%), Positives = 89/107 (83%)
Frame = +1
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY +VNDSC HQLVSHWL THAV+EPFVIAT+RHLS +HPI+KLL PHYR TM I
Sbjct: 361 WLLAKAYVIVNDSCYHQLVSHWLNTHAVIEPFVIATHRHLSFVHPIYKLLYPHYRDTMNI 540
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDL 193
N+ +R L+N G I+E TFL+ +Y MEMS+V+YK+WVF E+ LP DL
Sbjct: 541 NSLARGSLVNDGAIIEQTFLWGRYSMEMSSVLYKNWVFIEQSLPADL 681
>TC16035 similar to UP|O04919 (O04919) Lipoxygenase , partial (32%)
Length = 1035
Score = 129 bits (324), Expect = 5e-31
Identities = 53/87 (60%), Positives = 73/87 (82%)
Frame = +3
Query: 174 MSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWV 233
+S+ +YK+WVF E+ LP+DL RGMAV+D++SPHG+RL++EDYPYA+DGLEIW AI +WV
Sbjct: 3 LSSAIYKNWVFTEQALPSDLIKRGMAVEDENSPHGIRLVLEDYPYAADGLEIWGAISTWV 182
Query: 234 DEYVNFYYESDKDVKDDEELKAFWKEL 260
+YV YY SD VK+D EL+++WKE+
Sbjct: 183 QDYVTLYYVSDDAVKEDSELQSWWKEV 263
>AV424491
Length = 240
Score = 97.8 bits (242), Expect = 2e-21
Identities = 40/71 (56%), Positives = 58/71 (81%)
Frame = +2
Query: 190 PTDLKNRGMAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKD 249
P DL RGMA++D S+P+GLRL++EDYP+A DGLEIW AIK+WV +YV+ YY +D +++
Sbjct: 5 PADLIKRGMAIEDPSAPYGLRLVVEDYPFAVDGLEIWDAIKTWVLDYVSLYYSTDDEIQK 184
Query: 250 DEELKAFWKEL 260
D EL+ +WK++
Sbjct: 185 DTELQDWWKDV 217
>BP050172
Length = 594
Score = 91.3 bits (225), Expect = 2e-19
Identities = 39/59 (66%), Positives = 50/59 (84%)
Frame = +3
Query: 202 DDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
D +S HGLRLLIEDYPYA DGLEIW AI++WV +Y +FYY++D VK+D EL+++WKEL
Sbjct: 267 DPTSRHGLRLLIEDYPYAVDGLEIWFAIRTWVQDYXSFYYKTDDTVKEDAELQSWWKEL 443
>TC10343 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein
(Fibrillarin 2) (AtFib2), partial (85%)
Length = 1116
Score = 34.7 bits (78), Expect = 0.017
Identities = 17/32 (53%), Positives = 17/32 (53%)
Frame = +1
Query: 24 PPPRGRGGGGRPARGDDRGGDPADGKNRDESR 55
PPPRGRGGGG DRG DG R R
Sbjct: 37 PPPRGRGGGGGFRGRGDRGRGRGDGGGRGGGR 132
Score = 29.6 bits (65), Expect = 0.54
Identities = 19/46 (41%), Positives = 22/46 (47%), Gaps = 5/46 (10%)
Frame = +1
Query: 3 GQGERSERTPAPDGRGESGPSPPPRGRGGG-----GRPARGDDRGG 43
G+G+R GRG G P + RGGG GR RG RGG
Sbjct: 76 GRGDRGRGRGDGGGRG-GGRGTPFKARGGGRGGGGGRGGRGGGRGG 210
Score = 25.8 bits (55), Expect = 7.9
Identities = 20/53 (37%), Positives = 21/53 (38%), Gaps = 9/53 (16%)
Frame = +1
Query: 8 SERTPAPDGRGESGP----SPPPRGRG-----GGGRPARGDDRGGDPADGKNR 51
S+ P P GRG G RGRG GGGR RGG G R
Sbjct: 25 SKMVPPPRGRGGGGGFRGRGDRGRGRGDGGGRGGGRGTPFKARGGGRGGGGGR 183
>TC9669 similar to UP|Q9LQA7 (Q9LQA7) F4N2.10, partial (16%)
Length = 472
Score = 34.3 bits (77), Expect = 0.022
Identities = 23/78 (29%), Positives = 31/78 (39%), Gaps = 7/78 (8%)
Frame = +3
Query: 8 SERTPAPD---GRGESGPSPPPRGRGGGGR----PARGDDRGGDPADGKNRDESRARKHT 60
S ++P P G G S PP GGGG + +GG+ D S +
Sbjct: 234 SHKSPPPSPHHGSGGGHGSTPPSHGGGGGYYNPPSSPSPPKGGNCGSSPPHDPSTPTTPS 413
Query: 61 RTHNPTLPYPPPGPPNST 78
H P+ P+ P P ST
Sbjct: 414 TPHTPSTPHTPSTPTPST 467
>TC9801 homologue to UP|Q8LAC6 (Q8LAC6) Glycine rich protein-like, complete
Length = 981
Score = 33.1 bits (74), Expect = 0.049
Identities = 24/56 (42%), Positives = 27/56 (47%), Gaps = 5/56 (8%)
Frame = +2
Query: 7 RSERTPAPDGRGESGPSPPPRGRGG-----GGRPARGDDRGGDPADGKNRDESRAR 57
R++R P GRG RGRGG GGRPA+G RG D DG R R
Sbjct: 353 RTDRKPPGVGRG--------RGRGGREEGAGGRPAKGPGRGFD--DGAKGPGGRGR 490
Score = 25.8 bits (55), Expect = 7.9
Identities = 19/51 (37%), Positives = 23/51 (44%), Gaps = 1/51 (1%)
Frame = +2
Query: 2 GGQGERSERTPAPD-GRGESGPSPPPRGRGGGGRPARGDDRGGDPADGKNR 51
GG+ E + PA GRG + P GRG GG G GG G+ R
Sbjct: 398 GGREEGAGGRPAKGPGRGFDDGAKGPGGRGRGG---LGGKPGGSRGAGRGR 541
>BP066356
Length = 428
Score = 32.7 bits (73), Expect = 0.064
Identities = 14/35 (40%), Positives = 21/35 (60%)
Frame = +1
Query: 74 PPNSTAYHAGTFNWLLAKAYAVVNDSCCHQLVSHW 108
P ++T+Y + W + KA+ ND+ HQLV HW
Sbjct: 283 PMDATSY----WQWQIGKAHVSSNDAGVHQLVQHW 375
>TC8503 similar to GB|AAD30253.1|4835787|F25C20 ESTs gb|R65381 and
gb|T44635 come from this gene. {Arabidopsis thaliana;},
partial (26%)
Length = 1151
Score = 32.3 bits (72), Expect = 0.084
Identities = 27/81 (33%), Positives = 33/81 (40%), Gaps = 10/81 (12%)
Frame = -3
Query: 3 GQGERSERTPAPDGRGESGPSPPPRG-RGGGGRPARGDDRGGDPADGKNRDESRARKH-- 59
G G RS TP+PD + +GP P G R GGG G+ G+ S R+H
Sbjct: 429 GAGARSSPTPSPDHQ-PAGPRTPATGHRIGGGCSC---SNPGESGAGRRSRRSSHRRHRL 262
Query: 60 -------TRTHNPTLPYPPPG 73
R H YP PG
Sbjct: 261 APKRRHRRRHHRRWTRYPLPG 199
>BP084531
Length = 362
Score = 32.0 bits (71), Expect = 0.11
Identities = 14/25 (56%), Positives = 17/25 (68%)
Frame = +1
Query: 102 HQLVSHWLKTHAVVEPFVIATNRHL 126
H + H L THAV+EPF IA +R L
Sbjct: 286 HPFMYHRLHTHAVIEPFDIAASRQL 360
>TC8984 similar to UP|Q8LBA3 (Q8LBA3) Phosphoserine aminotransferase,
partial (25%)
Length = 563
Score = 32.0 bits (71), Expect = 0.11
Identities = 14/31 (45%), Positives = 16/31 (51%)
Frame = +1
Query: 16 GRGESGPSPPPRGRGGGGRPARGDDRGGDPA 46
G S P+PPP R RP+R G DPA
Sbjct: 241 GSSTSPPAPPPSRRTSSSRPSRSSTTGVDPA 333
>AU251519
Length = 412
Score = 31.6 bits (70), Expect = 0.14
Identities = 17/51 (33%), Positives = 21/51 (40%)
Frame = +2
Query: 15 DGRGESGPSPPPRGRGGGGRPARGDDRGGDPADGKNRDESRARKHTRTHNP 65
+ G + S PP RGG D GGD D + RA KH+ P
Sbjct: 200 EAAGRNRHSLPPSRRGGESDGGSDSDSGGDDDDSDDERSYRAGKHSAMQVP 352
>BP052983
Length = 443
Score = 31.6 bits (70), Expect = 0.14
Identities = 25/78 (32%), Positives = 30/78 (38%), Gaps = 4/78 (5%)
Frame = +1
Query: 11 TPAPDGRGESGPSPPPRGRGGGGRPARGDD---RGGDPADGKNRDESRARKHTRTHNPTL 67
TPA SGP+ P G A G+ D N D R+RKH P
Sbjct: 211 TPASYTLKSSGPTSPATGTSSRSPHAHGNSPLYSSPPKXDPGNPDTXRSRKHQYKLKP-- 384
Query: 68 PYPPPGP-PNSTAYHAGT 84
PP P P+ + H GT
Sbjct: 385 --PPTAPSPDPESIHYGT 432
>TC17500 UP|Q9ZPL9 (Q9ZPL9) Nodule-enhanced protein phosphatase type 2C,
complete
Length = 1325
Score = 31.2 bits (69), Expect = 0.19
Identities = 23/69 (33%), Positives = 28/69 (40%), Gaps = 11/69 (15%)
Frame = -3
Query: 22 PSPPPRGRGGGGRPARGDDR--GGDPADGKNRDESR---ARKHTRTHNPTLPYPP----- 71
P PPP R P + D G P RD + +R +RT +P P PP
Sbjct: 597 PPPPPHSRTSQPPPPQADTAAPGTPPPPELRRDRQKQRSSRTSSRTLSPRSPRPPSPSCS 418
Query: 72 -PGPPNSTA 79
P PN TA
Sbjct: 417 PPPKPNRTA 391
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.137 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,626,209
Number of Sequences: 28460
Number of extensions: 96291
Number of successful extensions: 1477
Number of sequences better than 10.0: 140
Number of HSP's better than 10.0 without gapping: 1332
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1433
length of query: 260
length of database: 4,897,600
effective HSP length: 88
effective length of query: 172
effective length of database: 2,393,120
effective search space: 411616640
effective search space used: 411616640
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 54 (25.4 bits)
Medicago: description of AC149580.1


