

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149577.7 - phase: 0
(173 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
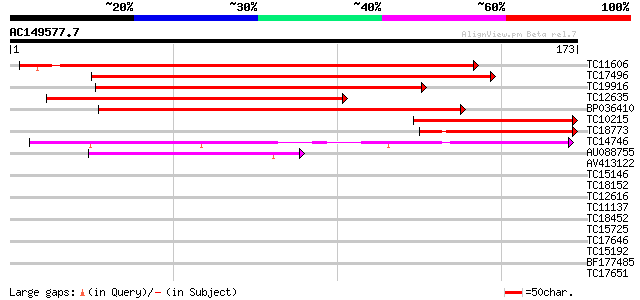
Score E
Sequences producing significant alignments: (bits) Value
TC11606 weakly similar to UP|Q9LW29 (Q9LW29) Transport inhibitor... 239 1e-64
TC17496 similar to PIR|T48087|T48087 transport inhibitor respons... 162 3e-41
TC19916 similar to PIR|T48087|T48087 transport inhibitor respons... 131 7e-32
TC12635 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor respon... 111 6e-26
BP036410 99 3e-22
TC10215 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor respon... 78 7e-16
TC18773 similar to PIR|T48087|T48087 transport inhibitor respons... 52 4e-08
TC14746 weakly similar to GB|AAG21976.1|10716947|AF263377 SKP1 i... 45 9e-06
AU088755 42 8e-05
AV413122 29 0.50
TC15146 similar to UP|AAR01764 (AAR01764) Expressed protein, par... 28 0.86
TC18152 27 1.9
TC12616 27 2.5
TC11137 similar to GB|CAD59563.1|27368811|OSA535041 PDR-like ABC... 26 3.3
TC18452 weakly similar to GB|AAO64875.1|29028992|BT005940 At5g27... 26 4.3
TC15725 similar to UP|MDHG_ARATH (Q9ZP05) Malate dehydrogenase, ... 26 4.3
TC17646 similar to UP|Q94A79 (Q94A79) AT3g49250/F2K15_110, parti... 25 7.3
TC15192 similar to UP|O80784 (O80784) Nodulin-like protein, part... 25 7.3
BF177485 25 9.5
TC17651 weakly similar to UP|Q8KBL5 (Q8KBL5) Oxidoreductase, sho... 25 9.5
>TC11606 weakly similar to UP|Q9LW29 (Q9LW29) Transport inhibitor
response-like protein, partial (18%)
Length = 892
Score = 239 bits (611), Expect = 1e-64
Identities = 120/141 (85%), Positives = 129/141 (91%), Gaps = 1/141 (0%)
Frame = +2
Query: 4 VECK-RKKESQGEKNNNMDSNSDFPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSR 62
+EC+ RKKES + N ++ S FPDEVLERVLGM+KSRKDRSSVSLVCKEWYNAERWSR
Sbjct: 473 MECRIRKKESP--EPNILNQASPFPDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSR 646
Query: 63 RNVFIGNCYAVSPEILTRRFPNIRSVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYP 122
R+VFIGNCY+VSPEILTRRFPNIRSVT+KGKPRFSDFNLVPANWGADI SWLVVFADKYP
Sbjct: 647 RSVFIGNCYSVSPEILTRRFPNIRSVTLKGKPRFSDFNLVPANWGADIRSWLVVFADKYP 826
Query: 123 FLEELRLKRMAVSDESLEFLA 143
LEELRLKRM VSDESLEFLA
Sbjct: 827 LLEELRLKRMTVSDESLEFLA 889
>TC17496 similar to PIR|T48087|T48087 transport inhibitor response protein
TIR1 [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (22%)
Length = 535
Score = 162 bits (410), Expect = 3e-41
Identities = 70/123 (56%), Positives = 92/123 (73%)
Frame = +3
Query: 26 FPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNI 85
FP+EVLE V ++ DR+++SLVCK WY ERW RR VF+GNCYAVSP I+ +RFP +
Sbjct: 165 FPEEVLEHVFSFIQVDTDRNAISLVCKSWYEIERWCRRKVFVGNCYAVSPMIVIKRFPEV 344
Query: 86 RSVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESLEFLAFS 145
RS+ +KGKP F+DFNLVP WG + +W+ + +P+LEE+RLKRM +SDESLE +A S
Sbjct: 345 RSIALKGKPHFADFNLVPEGWGGYVCTWIAAMSRAFPWLEEIRLKRMVISDESLELIAKS 524
Query: 146 FPN 148
F N
Sbjct: 525 FKN 533
>TC19916 similar to PIR|T48087|T48087 transport inhibitor response protein
TIR1 [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (16%)
Length = 446
Score = 131 bits (329), Expect = 7e-32
Identities = 58/101 (57%), Positives = 73/101 (71%)
Frame = +1
Query: 27 PDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNIR 86
P+EVLE V + + KDRSS+SLVC+ WY ER RRNVF+GNCYAVSPE++ +RFP +R
Sbjct: 142 PEEVLEHVFSFIDNDKDRSSISLVCRSWYEIERCCRRNVFVGNCYAVSPEMVVKRFPRVR 321
Query: 87 SVTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEEL 127
SVT+KGKP F+DFNLVP WG + W+ A P L E+
Sbjct: 322 SVTLKGKPHFADFNLVPDGWGGYVSPWIKAMAAACPSLLEI 444
>TC12635 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor response-like
protein, partial (14%)
Length = 696
Score = 111 bits (278), Expect = 6e-26
Identities = 49/92 (53%), Positives = 70/92 (75%)
Frame = -1
Query: 12 SQGEKNNNMDSNSDFPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCY 71
S+GE + + FPDEV+E V + S +DR+++SLVCK WY E+ +R+ VFIGNCY
Sbjct: 279 SEGE*EKGVKMMNYFPDEVIEHVFDYVVSHRDRNALSLVCKSWYRIEKSTRKRVFIGNCY 100
Query: 72 AVSPEILTRRFPNIRSVTMKGKPRFSDFNLVP 103
++SPE L +RFP++RS+T+KGKP F+DF+LVP
Sbjct: 99 SISPERLIQRFPSLRSLTLKGKPHFADFSLVP 4
>BP036410
Length = 550
Score = 99.4 bits (246), Expect = 3e-22
Identities = 46/112 (41%), Positives = 68/112 (60%)
Frame = +3
Query: 28 DEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNIRS 87
DEVL+ V+ + KDR +VS VCK WY + +R++V I CY +P L RRFP++ S
Sbjct: 207 DEVLDCVIPYIDDPKDRDAVSQVCKCWYELDSLTRKHVTIALCYTTTPARLRRRFPHLES 386
Query: 88 VTMKGKPRFSDFNLVPANWGADIHSWLVVFADKYPFLEELRLKRMAVSDESL 139
+ +KGKPR + FNL+P +WG + W++ + L+ L +RM V D L
Sbjct: 387 LKLKGKPRAAMFNLIPEDWGGHVTPWVLEINQYFDCLKSLHFRRMIVKDNDL 542
>TC10215 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor response-like
protein, partial (43%)
Length = 837
Score = 78.2 bits (191), Expect = 7e-16
Identities = 38/50 (76%), Positives = 44/50 (88%)
Frame = +3
Query: 124 LEELRLKRMAVSDESLEFLAFSFPNFKALSLLSCDGFSTDGLAAVATNCK 173
LEELRLKRM VSDESLE L+ SF NFK+L L+SC+GF+TDGLAAVA NC+
Sbjct: 54 LEELRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCR 203
>TC18773 similar to PIR|T48087|T48087 transport inhibitor response protein
TIR1 [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (50%)
Length = 927
Score = 52.4 bits (124), Expect = 4e-08
Identities = 24/48 (50%), Positives = 36/48 (75%)
Frame = +3
Query: 126 ELRLKRMAVSDESLEFLAFSFPNFKALSLLSCDGFSTDGLAAVATNCK 173
E+RL+ + D+ L+ +A SF NF+ L L+SC+GF+T GLAA+A NC+
Sbjct: 15 EIRLRGW-LYDDXLDLIAKSFKNFRVLVLISCEGFTTHGLAAIAANCR 155
>TC14746 weakly similar to GB|AAG21976.1|10716947|AF263377 SKP1 interacting
partner 1 {Arabidopsis thaliana;}, partial (62%)
Length = 719
Score = 44.7 bits (104), Expect = 9e-06
Identities = 47/169 (27%), Positives = 71/169 (41%), Gaps = 3/169 (1%)
Frame = +3
Query: 7 KRKKESQGEKNNNMDSN-SDFPDEVLERVLGMMKSRKDRSSVSLVCKEWYNA-ERWSRRN 64
+ K+E + E DS S+ E L +L + LVCK W++A + S +
Sbjct: 54 RNKEEMENEVEEGSDSEWSELTRECLINILSRLTVDDLWRGTMLVCKSWFSAFKEPSLHS 233
Query: 65 VFIGNCYAVSPEILTRRFPNIRSVTMKGKPRFSDFNLVPANWGADIHSWL-VVFADKYPF 123
VF + SP L R + P F A I S L V + F
Sbjct: 234 VFNLDPLFDSPRELPRWW----------SPEFE----------AKIDSMLRSVVQWTHIF 353
Query: 124 LEELRLKRMAVSDESLEFLAFSFPNFKALSLLSCDGFSTDGLAAVATNC 172
L ++R++ SD SL +A PN + LS+ SC + D ++ +A NC
Sbjct: 354 LTQIRIRHC--SDRSLALVAERCPNLEVLSIRSCPHVTDDSISRIAVNC 494
>AU088755
Length = 431
Score = 41.6 bits (96), Expect = 8e-05
Identities = 23/69 (33%), Positives = 37/69 (53%), Gaps = 3/69 (4%)
Frame = +2
Query: 25 DFPDEVLERVLGMMKSRKDRSSVSLVCKEWYNAERWSRRNVFIGNCYAVSPEILT---RR 81
+ P+ ++ +L +K DR+SVSLVCK YN + R ++ +G + E LT R
Sbjct: 47 NIPEHLVWEILSRIKKTSDRNSVSLVCKRLYNLDNGQRNSLKVGCGMDPADEALTCLCTR 226
Query: 82 FPNIRSVTM 90
F N+ V +
Sbjct: 227 FLNLSKVXI 253
>AV413122
Length = 399
Score = 28.9 bits (63), Expect = 0.50
Identities = 15/35 (42%), Positives = 20/35 (56%)
Frame = +2
Query: 118 ADKYPFLEELRLKRMAVSDESLEFLAFSFPNFKAL 152
A K+P LEEL + ++S E LE + S P K L
Sbjct: 248 AKKFPQLEELDISISSLSKEPLEVIGRSCPRLKTL 352
Score = 25.0 bits (53), Expect = 7.3
Identities = 17/55 (30%), Positives = 25/55 (44%), Gaps = 1/55 (1%)
Frame = +2
Query: 119 DKYPFLEELRLKRM-AVSDESLEFLAFSFPNFKALSLLSCDGFSTDGLAAVATNC 172
D L LRL ++SDE L +A FP + L +S S + L + +C
Sbjct: 173 DSTSHLRRLRLACCYSISDEGLSEIAKKFPQLEELD-ISISSLSKEPLEVIGRSC 334
>TC15146 similar to UP|AAR01764 (AAR01764) Expressed protein, partial (39%)
Length = 612
Score = 28.1 bits (61), Expect = 0.86
Identities = 16/55 (29%), Positives = 29/55 (52%), Gaps = 2/55 (3%)
Frame = -3
Query: 1 MNMVECKRKKESQGEK--NNNMDSNSDFPDEVLERVLGMMKSRKDRSSVSLVCKE 53
+N++ R SQ K +++ + DFPD++ ER+ G R+ ++ V L E
Sbjct: 472 LNVILLYRHSCSQPSKEGSSSCEGT*DFPDKLNERIKGCTMGRR*KAEVELATSE 308
>TC18152
Length = 647
Score = 26.9 bits (58), Expect = 1.9
Identities = 13/26 (50%), Positives = 14/26 (53%)
Frame = -2
Query: 5 ECKRKKESQGEKNNNMDSNSDFPDEV 30
EC R+ S GE N D N D DEV
Sbjct: 286 ECFRRDPSLGEVTYNDDDNGDTDDEV 209
>TC12616
Length = 434
Score = 26.6 bits (57), Expect = 2.5
Identities = 15/49 (30%), Positives = 24/49 (48%), Gaps = 1/49 (2%)
Frame = +3
Query: 8 RKKESQGEKNNNMDSN-SDFPDEVLERVLGMMKSRKDRSSVSLVCKEWY 55
R K ++G+++ N PD++ L + R D + LVCK WY
Sbjct: 183 RSKPARGDRSRNQSPLLPGLPDDLAIACL-IRVPRVDHRKLRLVCKRWY 326
>TC11137 similar to GB|CAD59563.1|27368811|OSA535041 PDR-like ABC
transporter {Oryza sativa (japonica cultivar-group);},
partial (15%)
Length = 786
Score = 26.2 bits (56), Expect = 3.3
Identities = 11/26 (42%), Positives = 17/26 (65%)
Frame = +1
Query: 95 RFSDFNLVPANWGADIHSWLVVFADK 120
R D+NLV ++ + SWLV+F+ K
Sbjct: 19 RSPDYNLVRFSFPCSLPSWLVLFSGK 96
>TC18452 weakly similar to GB|AAO64875.1|29028992|BT005940 At5g27920
{Arabidopsis thaliana;}, partial (11%)
Length = 698
Score = 25.8 bits (55), Expect = 4.3
Identities = 11/26 (42%), Positives = 16/26 (61%)
Frame = +2
Query: 148 NFKALSLLSCDGFSTDGLAAVATNCK 173
N K L + C ++ GLA++A NCK
Sbjct: 374 NLKTLEIRGCLLVTSIGLASIAMNCK 451
>TC15725 similar to UP|MDHG_ARATH (Q9ZP05) Malate dehydrogenase, glyoxysomal
precursor (mbNAD-MDH) , partial (39%)
Length = 586
Score = 25.8 bits (55), Expect = 4.3
Identities = 18/50 (36%), Positives = 26/50 (52%)
Frame = +1
Query: 48 SLVCKEWYNAERWSRRNVFIGNCYAVSPEILTRRFPNIRSVTMKGKPRFS 97
SL+C + + RWSRR++ G SPE + + + T KGK R S
Sbjct: 265 SLLCNQGTSRSRWSRRDISTG-----SPE*V*KDW------TGKGKERVS 381
>TC17646 similar to UP|Q94A79 (Q94A79) AT3g49250/F2K15_110, partial (12%)
Length = 611
Score = 25.0 bits (53), Expect = 7.3
Identities = 14/34 (41%), Positives = 18/34 (52%)
Frame = -1
Query: 112 SWLVVFADKYPFLEELRLKRMAVSDESLEFLAFS 145
+WLV K PFL + +S SLEF+ FS
Sbjct: 380 AWLV----KLPFLSSNNCCSLCISAASLEFIHFS 291
>TC15192 similar to UP|O80784 (O80784) Nodulin-like protein, partial (3%)
Length = 629
Score = 25.0 bits (53), Expect = 7.3
Identities = 8/21 (38%), Positives = 13/21 (61%)
Frame = -3
Query: 47 VSLVCKEWYNAERWSRRNVFI 67
+S CK W+N +RW + F+
Sbjct: 423 LSKYCKTWHNNQRWRYLHFFL 361
>BF177485
Length = 334
Score = 24.6 bits (52), Expect = 9.5
Identities = 12/31 (38%), Positives = 18/31 (57%)
Frame = -2
Query: 130 KRMAVSDESLEFLAFSFPNFKALSLLSCDGF 160
K +A+ +++F NF+A SLL DGF
Sbjct: 174 KHLALLRLRNRYVSFILQNFRATSLLDEDGF 82
>TC17651 weakly similar to UP|Q8KBL5 (Q8KBL5) Oxidoreductase, short-chain
dehydrogenase/reductase family, partial (12%)
Length = 599
Score = 24.6 bits (52), Expect = 9.5
Identities = 8/24 (33%), Positives = 16/24 (66%)
Frame = +1
Query: 79 TRRFPNIRSVTMKGKPRFSDFNLV 102
TRR PN ++++K +P +N++
Sbjct: 178 TRRMPNTTTLSLKSEPMLPPYNVL 249
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.134 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,136,910
Number of Sequences: 28460
Number of extensions: 38850
Number of successful extensions: 235
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 231
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 232
length of query: 173
length of database: 4,897,600
effective HSP length: 84
effective length of query: 89
effective length of database: 2,506,960
effective search space: 223119440
effective search space used: 223119440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 52 (24.6 bits)
Medicago: description of AC149577.7


