

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149574.3 + phase: 0
(489 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
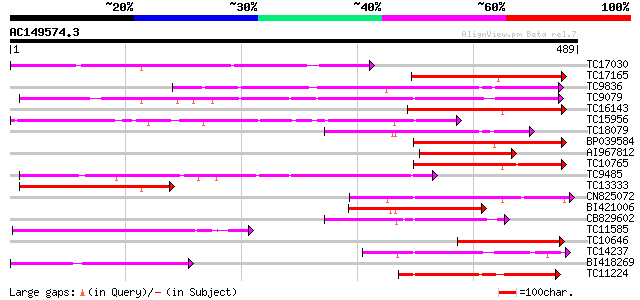
Score E
Sequences producing significant alignments: (bits) Value
TC17030 similar to UP|Q8S995 (Q8S995) Glucosyltransferase-14, pa... 215 1e-56
TC17165 similar to UP|Q9SMG6 (Q9SMG6) Betanidin-5-O-glucosyltran... 211 2e-55
TC9836 weakly similar to UP|Q9LXV0 (Q9LXV0) Glucosyltransferase-... 196 7e-51
TC9079 weakly similar to UP|Q8W4G1 (Q8W4G1) UDP-glucose glucosyl... 176 1e-44
TC16143 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltrans... 157 5e-39
TC15956 weakly similar to UP|LGT_CITUN (Q9MB73) Limonoid UDP-glu... 153 7e-38
TC18079 weakly similar to UP|UFO5_MANES (Q40287) Flavonol 3-O-gl... 143 6e-35
BP039584 142 9e-35
AI967812 141 2e-34
TC10765 homologue to UP|Q9ZWQ5 (Q9ZWQ5) UDP-glycose:flavonoid gl... 137 4e-33
TC9485 similar to UP|Q8S999 (Q8S999) Glucosyltransferase-10, par... 134 4e-32
TC13333 weakly similar to UP|AAR06917 (AAR06917) UDP-glycosyltra... 132 1e-31
CN825072 127 4e-30
BI421006 117 3e-27
CB829602 116 7e-27
TC11585 similar to UP|Q9ZWQ5 (Q9ZWQ5) UDP-glycose:flavonoid glyc... 113 8e-26
TC10646 weakly similar to UP|AAS55083 (AAS55083) UDP-glucose glu... 113 8e-26
TC14237 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltra... 108 3e-24
BI418269 108 3e-24
TC11224 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferase... 103 5e-23
>TC17030 similar to UP|Q8S995 (Q8S995) Glucosyltransferase-14, partial (51%)
Length = 1098
Score = 215 bits (548), Expect = 1e-56
Identities = 122/317 (38%), Positives = 179/317 (55%), Gaps = 3/317 (0%)
Frame = +1
Query: 1 MDSQSNPLHILVFPFMGHGHTIPTIDMAKLFASK-GVRVTIVTTPLNKPPISKALEQSKI 59
M SQ++ +H ++FP M GH IP +D+A + + + V VT++TTP N + +
Sbjct: 187 MASQASQIHFVLFPLMSQGHMIPMMDIATILSQQQDVTVTVITTPHNASRFTHTTKSR-- 360
Query: 60 HFNNIDIQTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELL--LQQKP 117
+ I I ++FP E GLPEGCEN+D +PS+ FF A LQ+P E+L L P
Sbjct: 361 --SQIRILELEFPHNETGLPEGCENLDMLPSLGTALTFFNAASNLQKPVEQLFNELTPPP 534
Query: 118 HCVVADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITD 177
+C+++DM P+ AAKF IPRI F G S F+L + +++ +NV+S+T+ F +
Sbjct: 535 NCIISDMCLPYTARIAAKFNIPRISFLGQSCFTLFCLYNIGRHKVRQNVTSETEYFVLPG 714
Query: 178 LPGNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYR 237
+P I+MT+ Q+P + Q+ + E +E SYGV++NSF ELE YA Y+
Sbjct: 715 VPDKIEMTKAQIPG--PSGARMDQNRIAFYAETAAAEAASYGVVMNSFEELETEYAKGYK 888
Query: 238 EVLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMT 297
+V K W IGP S+ N NK D+H C+KWLD + SV+Y C GSM
Sbjct: 889 KVKNGKVWCIGPVSLSNNNK---------VSIDEDEHYCMKWLDLQKSKSVIYACLGSMC 1041
Query: 298 HFLNSQLKEIAMGLEAS 314
+ QL E+ +GLEAS
Sbjct: 1042NLTPLQLIELGLGLEAS 1092
>TC17165 similar to UP|Q9SMG6 (Q9SMG6) Betanidin-5-O-glucosyltransferase,
partial (24%)
Length = 546
Score = 211 bits (537), Expect = 2e-55
Identities = 105/136 (77%), Positives = 116/136 (85%), Gaps = 2/136 (1%)
Frame = +1
Query: 347 IIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLK 406
IIRGW+PQV+ILEHEAIGAFVTHCGWNS LEGV AGVPM+TWP+AAEQFYNEKLVTEVLK
Sbjct: 1 IIRGWAPQVLILEHEAIGAFVTHCGWNSTLEGVAAGVPMVTWPIAAEQFYNEKLVTEVLK 180
Query: 407 TGVPVGVKKWVMKV--GDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDG 464
GVPVG KKW V GD V+ DAVEK VKR+MEGEEA EMRN+ K+L++ A+ AVEE G
Sbjct: 181 IGVPVGAKKWGRMVLEGDGVKSDAVEKGVKRIMEGEEAEEMRNRVKVLSQQAQWAVEEGG 360
Query: 465 SSYSQLNALIEELRSL 480
SSYS LNALIEEL L
Sbjct: 361 SSYSDLNALIEELGML 408
>TC9836 weakly similar to UP|Q9LXV0 (Q9LXV0) Glucosyltransferase-like
protein, partial (55%)
Length = 1286
Score = 196 bits (498), Expect = 7e-51
Identities = 129/346 (37%), Positives = 183/346 (52%), Gaps = 9/346 (2%)
Frame = +1
Query: 141 IVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLPGNIKMTRLQLPNTLTENDPIS 200
++F G+S + L + P++ V D+D F + D P ++ R QLP+ + E D
Sbjct: 1 VIFSGSSGYGLACYFSLWVNLPHRRV--DSDHFPLPDFPEAGQIHRTQLPSNIAEAD--G 168
Query: 201 QSFAKLFEEIKDSE-VRSYGVIVNSFYELENVYADYYREVLGIKEWHIGPFSIHNRNKEE 259
+ LF+ S V S G + N+ + +++ Y L W IGP + +
Sbjct: 169 EDAWSLFQISNISNWVNSDGFLFNTVADFDSMGLRYVARKLNRPAWAIGPVLLSTGSGS- 345
Query: 260 EIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLNSQLKEIAMGLEASGHNFI 319
RGK I C KWLDTK NSV+++ FGSM SQ+ ++A L+ SG NFI
Sbjct: 346 -----RGKGGGISPELCRKWLDTKPSNSVLFVSFGSMNTISASQMMQLATALDRSGRNFI 510
Query: 320 WVVR-------TQTEDGDEWLPEGFEERTEGKGLIIRGWSPQVMILEHEAIGAFVTHCGW 372
WVVR +EWLPEGF R KGL++ W+PQV IL H A+ AF++HCGW
Sbjct: 511 WVVRPPIGFDINSEFRAEEWLPEGFLGRVAKKGLVVHDWAPQVEILSHGAVSAFLSHCGW 690
Query: 373 NSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVKKWVMKVGDNVEWDAVEKA 432
NSVLE + GVP++ WP+AAEQF+N K++ E L GV V V + K + D VEK
Sbjct: 691 NSVLESLSHGVPILGWPMAAEQFFNCKMLEEEL--GVCVEVARG--KRCEVRHEDLVEKI 858
Query: 433 VKRVMEGEEAYEMRNKAKMLAEMAKKAV-EEDGSSYSQLNALIEEL 477
+ E E ++R A + EM + AV +EDG S + A+ E L
Sbjct: 859 ELVMNEAESGVKIRKNAGNIREMIRDAVRDEDGYKGSSVRAIDEFL 996
>TC9079 weakly similar to UP|Q8W4G1 (Q8W4G1) UDP-glucose
glucosyltransferase, partial (65%)
Length = 1585
Score = 176 bits (445), Expect = 1e-44
Identities = 136/490 (27%), Positives = 223/490 (44%), Gaps = 21/490 (4%)
Frame = +1
Query: 9 HILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIH-FNNIDIQ 67
H ++ P GH P +AKL +G +T V T N + K+ + + + +
Sbjct: 19 HAVLTPSPLQGHINPLFQLAKLLHLRGFHITFVHTEYNHKRLIKSRGPNALDGLPDFVFE 198
Query: 68 TIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELL----------LQQKP 117
TI P+G E+ D S + PF++LL L
Sbjct: 199 TI---------PDGLEDGDGEVSQDLYSLCDSIRNKCHLPFQDLLAKLHRSASAGLVPPV 351
Query: 118 HCVVADMFFPWATDSAAKFGIPRIVF---HGTSFFSLCASQCM--KKYQPYKNVSSDTDL 172
C+V+D+ + +A + +P ++ ++F S Q + K P K+ S T+
Sbjct: 352 TCLVSDLLMTFTIQAAQQLQLPILLLCPASASTFMSFLHFQTLLDKGVIPLKDESYLTNG 531
Query: 173 F---EITDLPGNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELE 229
+ ++ +PG LP+ + DP + + E+ D R+ ++ N+F ELE
Sbjct: 532 YLDTKVDWIPGMQNFRLKDLPDFIRTTDP-NDIMLEFMVEVADRAKRASAIVFNTFNELE 708
Query: 230 NVYADYYREVLGIKEWHIGPF-SIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSV 288
+L + IGPF S N+ + ++ S G + +CL+WL++K SV
Sbjct: 709 RDVLSALSTMLP-SLYPIGPFPSFLNQAPQNQLASL-GSNLWKEDTKCLQWLESKEPGSV 882
Query: 289 VYMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQ-TEDGDEWLPEGFEERTEGKGLI 347
VY+ FGS+T QL E A GL F+W++R DG L F + T +GLI
Sbjct: 883 VYVNFGSITVMSPEQLLEFAWGLANGKKPFLWIIRPDLVADGSAILSPKFVDETSNRGLI 1062
Query: 348 IRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKT 407
W PQ +L H ++G F+THCGWNS +E + AGVPM+ WP A+Q N + +
Sbjct: 1063A-SWCPQEKVLNHPSVGGFLTHCGWNSTIESICAGVPMLCWPFVADQPTNCRYICNEWDI 1239
Query: 408 GVPVGVKKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSY 467
G+ +V NV+ + VE + +M G++ +MR + L + A++ G SY
Sbjct: 1240GI---------EVDTNVKREEVENLIHELMVGDKGKKMRQRTMELKKKAEEDTRPGGCSY 1392
Query: 468 SQLNALIEEL 477
L+ LI+E+
Sbjct: 1393MNLDKLIKEV 1422
>TC16143 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltransferase,
partial (30%)
Length = 604
Score = 157 bits (396), Expect = 5e-39
Identities = 79/142 (55%), Positives = 104/142 (72%), Gaps = 5/142 (3%)
Frame = +1
Query: 344 KGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTE 403
KG+IIRGW PQV+IL H A+GAFVTHCGWNS +E V AGVPMITWPV EQFYNEKL+TE
Sbjct: 7 KGMIIRGWVPQVVILGHRAVGAFVTHCGWNSTVEAVSAGVPMITWPVVGEQFYNEKLITE 186
Query: 404 VLKTGVPVGVKKWVMKVGDN----VEWDAVEKAVKRVME-GEEAYEMRNKAKMLAEMAKK 458
V GV VG ++ + D V +++EKAV+R+M+ G+E ++R +A+ E A++
Sbjct: 187 VRGIGVEVGAEECCLMGFDKREKLVSRESIEKAVRRLMDGGDEGEQIRRRAQEYGEKARQ 366
Query: 459 AVEEDGSSYSQLNALIEELRSL 480
AVEE GSS+ L ALI++L+ L
Sbjct: 367 AVEEGGSSHKNLTALIDDLKRL 432
>TC15956 weakly similar to UP|LGT_CITUN (Q9MB73) Limonoid
UDP-glucosyltransferase (Limonoid glucosyltransferase)
(Limonoid GTase) (LGTase) , partial (65%)
Length = 1169
Score = 153 bits (386), Expect = 7e-38
Identities = 116/405 (28%), Positives = 200/405 (48%), Gaps = 16/405 (3%)
Frame = +2
Query: 1 MDSQSNPLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIH 60
M S+S PLHIL+ F GH P + +AK A+KG VT+ TT + A +
Sbjct: 17 MGSES-PLHILLVSFPAQGHINPLLRLAKCLAAKGSSVTLATTQTAGKSMRTATNITDKA 193
Query: 61 FNNIDIQTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKPH-- 118
I ++KF + GL +++ S+ + + L+ + +++L K H
Sbjct: 194 PTPISDGSLKFEFFDDGLGPDDDSIRGNLSL-----YLQQLELVGR--QQILKMIKEHAD 352
Query: 119 ------CVVADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKN----VSS 168
C++ + F PW D A + GIP ++ + +S Y Y + S
Sbjct: 353 SNRQISCIINNPFLPWVVDVAEEQGIP------SALLWIQSSAVFTAYYSYFHNLVTFPS 514
Query: 169 DTDLFEITDLPG-NIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYE 227
D LP +I + ++P+ L + + E+ K+ +++ ++V+++ E
Sbjct: 515 KEDPLADVQLPSTSIVLKHCEIPDFLHPSCTYPFLGTLILEQFKNLN-KTFCILVDTYEE 691
Query: 228 LENVYADYYREVLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINS 287
LE+ + DY + I+ +GP ++ K I RG D +C++WL++K +S
Sbjct: 692 LEHDFIDYLSKTFLIRP--VGPLFKSSQAKSAAI---RGDFMKSD--DCIEWLNSKPQSS 850
Query: 288 VVYMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGD---EWLPEGFEERTEGK 344
VVY+ FGS+ + Q+ EIA GL+ S +F+WV++ E LP GF E T +
Sbjct: 851 VVYVSFGSIVYLPQEQVDEIAFGLKKSQVSFLWVMKPPPEVTGLQPHVLPYGFLEETGER 1030
Query: 345 GLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWP 389
G +++ WSPQ +L H ++ F+THCGWNS +E + +GVP++T+P
Sbjct: 1031GRVVQ-WSPQEEVLAHPSVSCFLTHCGWNSSMEALTSGVPVLTFP 1162
>TC18079 weakly similar to UP|UFO5_MANES (Q40287) Flavonol
3-O-glucosyltransferase 5 (UDP-glucose flavonoid
3-O-glucosyltransferase 5) , partial (33%)
Length = 889
Score = 143 bits (361), Expect = 6e-35
Identities = 85/194 (43%), Positives = 112/194 (56%), Gaps = 13/194 (6%)
Frame = +3
Query: 272 DKHECLKWLDTKNINSVVYMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQTED--- 328
D+ E L+WLD + SV+Y+ FGS Q+ EIA+GLE S H F+WVVR E
Sbjct: 48 DQDEILRWLDKQPAGSVIYVSFGSGGTMPQGQMTEIALGLELSQHRFVWVVRPPNEADAS 227
Query: 329 ------GDE----WLPEGFEERTEGKGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEG 378
G+E +LPEGF RT G+++ W+PQ IL H A G FVT CGWNSVLE
Sbjct: 228 ATFFGFGNEMALNYLPEGFMNRTREFGVVVPMWAPQAEILGHPATGGFVTPCGWNSVLES 407
Query: 379 VVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVKKWVMKVGDNVEWDAVEKAVKRVME 438
V+ GVPM+ WP+ +EQ N +++E L GV V VK + G V D V V+RVM
Sbjct: 408 VLNGVPMVAWPLYSEQKMNAYMLSEEL--GVAVRVK---VAEGGVVCRDQVVDMVRRVMV 572
Query: 439 GEEAYEMRNKAKML 452
EE M+++ + L
Sbjct: 573 SEEGMGMKDRVREL 614
>BP039584
Length = 545
Score = 142 bits (359), Expect = 9e-35
Identities = 71/136 (52%), Positives = 99/136 (72%), Gaps = 4/136 (2%)
Frame = -3
Query: 349 RGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTG 408
RGW+PQ++ILEH AIG VTHCGWN+ LE V+AG+PM T P+ AEQFYNEKL+ +VL G
Sbjct: 543 RGWAPQLLILEHPAIGGVVTHCGWNTTLESVIAGLPMATMPLFAEQFYNEKLLVDVLGVG 364
Query: 409 VPVGVKKW--VMKVGDN-VEWDAVEKAVKRVM-EGEEAYEMRNKAKMLAEMAKKAVEEDG 464
V +G+KKW VGD V+ + + KA+ +M GEEA EMR + + L++ AKK ++ G
Sbjct: 363 VSIGMKKWRNWNVVGDEIVKRENIVKAISLLMGGGEEALEMRRRVRELSDAAKKTIQPGG 184
Query: 465 SSYSQLNALIEELRSL 480
SSY+++ L +EL++L
Sbjct: 183 SSYNKVKGLFDELKAL 136
>AI967812
Length = 274
Score = 141 bits (356), Expect = 2e-34
Identities = 65/85 (76%), Positives = 75/85 (87%), Gaps = 1/85 (1%)
Frame = +2
Query: 354 QVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGV 413
QV+ILEHEAIGAFVTHCGWNS LEGV AGVPM+TWP++AEQFYNEKLVT VLK GVPVGV
Sbjct: 17 QVLILEHEAIGAFVTHCGWNSTLEGVSAGVPMVTWPLSAEQFYNEKLVTHVLKIGVPVGV 196
Query: 414 KKWVMKVGD-NVEWDAVEKAVKRVM 437
KKW M GD ++WDA+ K++KR+M
Sbjct: 197 KKWTMFTGDGTIKWDAMVKSLKRIM 271
>TC10765 homologue to UP|Q9ZWQ5 (Q9ZWQ5) UDP-glycose:flavonoid
glycosyltransferase, partial (19%)
Length = 542
Score = 137 bits (345), Expect = 4e-33
Identities = 70/136 (51%), Positives = 95/136 (69%), Gaps = 4/136 (2%)
Frame = +2
Query: 349 RGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTG 408
+GW+PQ +IL H A G F+THCGWN+V+E + AGVPMIT P ++Q+YNEKL+TEV G
Sbjct: 2 KGWAPQPLILNHLATGGFLTHCGWNAVVEAISAGVPMITMPGFSDQYYNEKLITEVHGFG 181
Query: 409 VPVGVKKWVMKVGDN----VEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDG 464
V VG +W + D V + +EKAVK +M GEE E+R+KAK + + A KAV++ G
Sbjct: 182 VEVGAAEWSISPYDGKKKVVSGERIEKAVKSLM-GEEGAEIRSKAKEVQDKAWKAVQQGG 358
Query: 465 SSYSQLNALIEELRSL 480
SSY+ L LI+ LR+L
Sbjct: 359 SSYNSLTVLIDHLRTL 406
>TC9485 similar to UP|Q8S999 (Q8S999) Glucosyltransferase-10, partial (75%)
Length = 1195
Score = 134 bits (336), Expect = 4e-32
Identities = 111/378 (29%), Positives = 175/378 (45%), Gaps = 17/378 (4%)
Frame = +1
Query: 9 HILVFPFMGHGHTIPTIDMAKLFASKG-VRVTIVTTPLNKPPISKALEQSKIHFNNIDIQ 67
H++ P+ GH P + +AKL KG VT V T N + K+ ++ +
Sbjct: 91 HVVCIPYPAQGHINPMLKLAKLLHFKGGFHVTFVNTEYNHKRLLKSRGPDSLN----GLP 258
Query: 68 TIKFPCVEAGLPEGCENV-DSIPS--VSFVPAFFAAIRLLQQPFEELLLQQKP-HCVVAD 123
+ +F + GLPE +V IPS +S + L ++ P C+V+D
Sbjct: 259 SFRFETIPDGLPETDVDVTQDIPSLCISTRKTCLPHFKKLLSKLNDVSSDVPPVTCIVSD 438
Query: 124 MFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQ--------PYKNVSSDTDLFEI 175
+ D+A + IP ++F TS C +Y+ P K+ S T+ +
Sbjct: 439 GCMSFTLDAAIELNIPEVLFWTTS---ACGFMGYVQYRELIEKGIIPLKDSSDITNGYLE 609
Query: 176 TD---LPGNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVY 232
T LPG + LP+ L DP + L E + + +++ +I+N+F LE+
Sbjct: 610 TTIEWLPGMKNIRLKDLPSFLRTTDPNDKMLDFLTGECQRA-LKASAIILNTFDALEHDV 786
Query: 233 ADYYREVLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMC 292
+ + +L + IGP + ++ ++ + G + ECLKWLDTK NSVVY+
Sbjct: 787 LEAFSSILP-PVYSIGPLHLLIKDVTDKNLNSLGSNLWKEDSECLKWLDTKEPNSVVYVN 963
Query: 293 FGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGDE-WLPEGFEERTEGKGLIIRGW 351
FGS+T + Q+ E A GL S F+WV+R G LPE F T +G + W
Sbjct: 964 FGSITVMTSEQMVEFAWGLANSNKTFLWVIRPDLVAGKHAVLPEEFVAATNDRGR-LSSW 1140
Query: 352 SPQVMILEHEAIGAFVTH 369
+PQ +L H AIG F+TH
Sbjct: 1141TPQEDVLTHPAIGGFLTH 1194
>TC13333 weakly similar to UP|AAR06917 (AAR06917) UDP-glycosyltransferase
73E1, partial (10%)
Length = 419
Score = 132 bits (333), Expect = 1e-31
Identities = 62/136 (45%), Positives = 86/136 (62%), Gaps = 2/136 (1%)
Frame = +3
Query: 9 HILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFNNIDIQT 68
H L+ PFM H IP +AKL A+ G+ VTIV TPLN + ++Q+K I +
Sbjct: 12 HFLLVPFMSQSHLIPFTQLAKLLAANGITVTIVLTPLNATRFNMVIDQAKASNLKIHFKV 191
Query: 69 IKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELL--LQQKPHCVVADMFF 126
+ FPC EAGLPEGCEN+DS+ S P FFAA +L+QP E+ L L+ P C+++D+
Sbjct: 192 LPFPCKEAGLPEGCENMDSVTSPQHQPLFFAACNMLKQPLEKWLSELETVPTCIISDICL 371
Query: 127 PWATDSAAKFGIPRIV 142
PW + +A KF IPR++
Sbjct: 372 PWTSSTATKFNIPRVI 419
>CN825072
Length = 712
Score = 127 bits (319), Expect = 4e-30
Identities = 79/215 (36%), Positives = 116/215 (53%), Gaps = 21/215 (9%)
Frame = -3
Query: 294 GSMTHFLNSQLKEIAMGLEASGHNFIWVVRT-------------QTEDGDEWLPEGFEER 340
GS F +Q+ EIA +E SG F+W +R +D LPEGF +R
Sbjct: 710 GSRGSFDRAQITEIARAVENSGVRFVWSLRKPXLKGSMVGPSDYSVDDLASVLPEGFLDR 531
Query: 341 TEGKGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKL 400
T G G +I GW+PQ +L H A G FV+HCGWNS LE + GVP+ TWP+ AEQ N +
Sbjct: 530 TTGIGRVI-GWAPQTRVLTHPATGGFVSHCGWNSTLESIYFGVPIATWPLYAEQQTNAFV 354
Query: 401 VTEVLKTGVPVGVKKWV-MKVGDN--VEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAK 457
+ LK V + + V + G N + D +E ++ V++ + E+R + K ++E ++
Sbjct: 353 LVRELKIAVEISLDYRVEINGGPNYLLTADKIEGGIRSVLDKDG--EVRKRVKEMSEKSR 180
Query: 458 KAVEEDGSSYSQLNALIEEL-----RSLSHHQHIS 487
K + E G SYS L+ LI+ LS+H H+S
Sbjct: 179 KTLLEGGCSYSYLDRLIDYFMHQV*NHLSNHTHVS 75
>BI421006
Length = 519
Score = 117 bits (294), Expect = 3e-27
Identities = 57/127 (44%), Positives = 82/127 (63%), Gaps = 8/127 (6%)
Frame = +2
Query: 293 FGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQTE----DGDE----WLPEGFEERTEGK 344
F + F+ Q+++IA GLE S FIWV+R + DGD+ LP+GFE+R EG
Sbjct: 101 FWFFSTFIEEQIEQIANGLEQSKQKFIWVLRDADKGDIFDGDKVKERGLPKGFEKRVEGM 280
Query: 345 GLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEV 404
GL++R W+PQ+ IL H + G F++HCGWNS +E + GVP+ WP+ ++Q N L+TEV
Sbjct: 281 GLVVRDWAPQLEILSHPSTGGFMSHCGWNSCIESMSMGVPIAAWPMHSDQPRNTVLITEV 460
Query: 405 LKTGVPV 411
LK + V
Sbjct: 461 LKVALVV 481
>CB829602
Length = 542
Score = 116 bits (291), Expect = 7e-27
Identities = 62/162 (38%), Positives = 96/162 (58%), Gaps = 2/162 (1%)
Frame = +1
Query: 272 DKHECLKWLDTKNINSVVYMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGDE 331
++ ECLKWLD++ +SV+Y+ FGS+ + QL E+A G+ S FIWV+R +G+
Sbjct: 82 EEPECLKWLDSQEPSSVLYVNFGSVINMTPQQLVELAWGIANSKKKFIWVIRPDLVEGEA 261
Query: 332 W--LPEGFEERTEGKGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWP 389
LPE E T+ +G+++ W PQ +L+H A+G F+THCGWNS +E + +GVP+I P
Sbjct: 262 SIVLPEIVAE-TKDRGIML-SWCPQEQVLKHSALGGFLTHCGWNSTIESISSGVPLICSP 435
Query: 390 VAAEQFYNEKLVTEVLKTGVPVGVKKWVMKVGDNVEWDAVEK 431
+QF N + + G+ + D+V+ D VEK
Sbjct: 436 FFNDQFVNSRYICSEWDFGMEMN--------SDDVKRDEVEK 537
>TC11585 similar to UP|Q9ZWQ5 (Q9ZWQ5) UDP-glycose:flavonoid
glycosyltransferase, partial (40%)
Length = 609
Score = 113 bits (282), Expect = 8e-26
Identities = 67/208 (32%), Positives = 102/208 (48%)
Frame = +1
Query: 3 SQSNPLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFN 62
S+ L + PF GH IP + +A+L AS+G VTI+TTP N K +E+ +
Sbjct: 13 SEMETLKVYFVPFFAQGHLIPLVHLARLVASRGEHVTIITTPANAQLFDKTIEEDRACGY 192
Query: 63 NIDIQTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKPHCVVA 122
+I + +KFP + GLP G EN+ + A L+Q E + Q P ++
Sbjct: 193 HIRVHLVKFPSTQVGLPGGVENLFAASDNLTAGKIHMAAHLIQPEIEGFMKQSPPDVLIP 372
Query: 123 DMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLPGNI 182
D+ F W+ SA GIPR++F+ S F +C + +K + P SD+ + I PG
Sbjct: 373 DIMFTWSEASAKSLGIPRLIFNPISIFDVCMIEAIKSH-PEAFTCSDSGPYHI---PG-- 534
Query: 183 KMTRLQLPNTLTENDPISQSFAKLFEEI 210
LP++LT S FA+L E +
Sbjct: 535 ------LPHSLTLPIKPSPGFARLTESL 600
>TC10646 weakly similar to UP|AAS55083 (AAS55083) UDP-glucose
glucosyltransferase, partial (8%)
Length = 501
Score = 113 bits (282), Expect = 8e-26
Identities = 55/93 (59%), Positives = 74/93 (79%), Gaps = 1/93 (1%)
Frame = +1
Query: 387 TWPVAAEQFYNEKLVTEVLKTGVPVGVKKWVMKVG-DNVEWDAVEKAVKRVMEGEEAYEM 445
TWPV+AEQFYNEKLVT++L+ GVPVGVKKW VG D++ AVE+A+ R+M EEA
Sbjct: 1 TWPVSAEQFYNEKLVTDILEIGVPVGVKKWARVVGDDSITSSAVERAINRIMVQEEAESF 180
Query: 446 RNKAKMLAEMAKKAVEEDGSSYSQLNALIEELR 478
RN+A LA++A+ AV+++GSS+S L ALI++LR
Sbjct: 181 RNRAHKLAQVARTAVQDNGSSHSHLTALIQQLR 279
>TC14237 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltransferase
85A8, partial (27%)
Length = 999
Score = 108 bits (269), Expect = 3e-24
Identities = 68/185 (36%), Positives = 102/185 (54%), Gaps = 6/185 (3%)
Frame = +2
Query: 305 KEIAM-GLEASGHNFIWVVRTQTEDGDEWL--PEGFEERTEGKGLIIRGWSPQVMILEHE 361
KEI + G+ S F+W++R G++ + P+ F + +G+G I W Q +L H
Sbjct: 176 KEIWLWGIANSKVPFLWILRPDVVMGEDSINVPQEFLDEIKGRGYIA-SWCFQEEVLSHP 352
Query: 362 AIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVKKWVMKVG 421
+IGAF+THCGWNS +EG+ AG+P+I WP +EQ N + + G M+V
Sbjct: 353 SIGAFLTHCGWNSTIEGISAGLPLICWPFFSEQHTNSRYACTTWEIG---------MEVN 505
Query: 422 DNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEE---DGSSYSQLNALIEELR 478
+V+ D + V +M+GE+ EMR K E KKA+E GSSY+ + LI+E
Sbjct: 506 HDVKRDEITTLVNEMMKGEKGKEMRKKG---LEWKKKAIEATGLGGSSYNDFHKLIKE-- 670
Query: 479 SLSHH 483
SL H+
Sbjct: 671 SLHHN 685
>BI418269
Length = 520
Score = 108 bits (269), Expect = 3e-24
Identities = 55/158 (34%), Positives = 84/158 (52%)
Frame = +2
Query: 1 MDSQSNPLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIH 60
++S+ PL + P++ GH IP D+A LFAS+G VTI+TTP N I +L
Sbjct: 56 VESEQRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLP----- 220
Query: 61 FNNIDIQTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKPHCV 120
N+ + T+ FP + GLP+G E+ S + I+LL++P + + CV
Sbjct: 221 --NLHLHTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCV 394
Query: 121 VADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMK 158
VAD FPW D + IPR+ F+ + F+LCA + +
Sbjct: 395 VADYCFPWIDDLTTELHIPRLSFNPSPLFALCAMRAKR 508
>TC11224 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13
(Fragment), partial (19%)
Length = 596
Score = 103 bits (258), Expect = 5e-23
Identities = 61/140 (43%), Positives = 85/140 (60%)
Frame = +3
Query: 336 GFEERTEGKGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQF 395
GF ERT+ +G ++ W+PQ+ IL+H ++G F+TH GWNSVLE +V GVPMI P +Q
Sbjct: 6 GFLERTKSQGKVV-SWAPQMEILKHASVGVFLTHGGWNSVLECIVGGVPMIGRPFFGDQR 182
Query: 396 YNEKLVTEVLKTGVPVGVKKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEM 455
N ++ ++L GV VG++ V+ + + K +K M GEE MR K L EM
Sbjct: 183 INIWMLAKLL--GVGVGLQNGVLAK------ETILKTLKSTMTGEEGKVMRQKMAELKEM 338
Query: 456 AKKAVEEDGSSYSQLNALIE 475
A KAVE DGSS L L++
Sbjct: 339 AWKAVEPDGSSTKNLCTLMQ 398
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,822,875
Number of Sequences: 28460
Number of extensions: 123519
Number of successful extensions: 697
Number of sequences better than 10.0: 111
Number of HSP's better than 10.0 without gapping: 643
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 650
length of query: 489
length of database: 4,897,600
effective HSP length: 94
effective length of query: 395
effective length of database: 2,222,360
effective search space: 877832200
effective search space used: 877832200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC149574.3


