

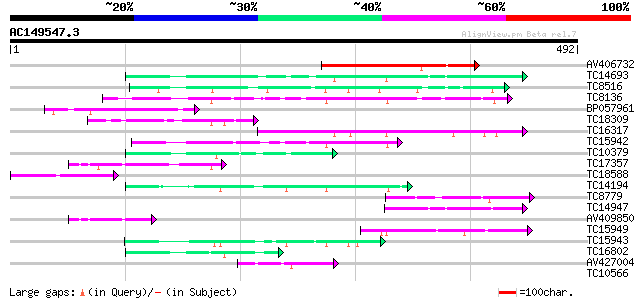
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149547.3 + phase: 0
(492 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV406732 149 8e-37
TC14693 similar to GB|BAB01116.1|11994113|AB026658 CND41, chloro... 95 2e-20
TC8516 similar to UP|AAP72988 (AAP72988) CDR1, partial (27%) 91 4e-19
TC8136 similar to GB|AAB60729.1|2160166|F21M12 F21M12.13 {Arabid... 90 1e-18
BP057961 76 1e-14
TC18309 similar to UP|Q9FG60 (Q9FG60) Aspartyl protease-like, pa... 75 2e-14
TC16317 similar to UP|Q8W4C5 (Q8W4C5) Nucellin-like protein, par... 72 2e-13
TC15942 similar to UP|Q9FL43 (Q9FL43) Nucleoid DNA-binding-like ... 68 4e-12
TC10379 weakly similar to UP|Q8H9F4 (Q8H9F4) 41 kDa chloroplast ... 67 7e-12
TC17357 similar to UP|Q9FFC3 (Q9FFC3) Protease-like protein, par... 64 4e-11
TC18588 61 5e-10
TC14194 weakly similar to UP|Q8GT67 (Q8GT67) Xyloglucan-specific... 58 4e-09
TC8779 similar to UP|Q8L934 (Q8L934) Nucleoid DNA-binding-like p... 52 2e-07
TC14947 similar to GB|AAP31963.1|30387595|BT006619 At1g01300 {Ar... 50 8e-07
AV409850 47 7e-06
TC15949 similar to UP|Q9LTW4 (Q9LTW4) Chloroplast nucleoid DNA b... 47 9e-06
TC15943 similar to UP|Q8GT67 (Q8GT67) Xyloglucan-specific fungal... 45 4e-05
TC16802 similar to GB|AAP31963.1|30387595|BT006619 At1g01300 {Ar... 44 5e-05
AV427004 42 3e-04
TC10566 similar to UP|Q9LI73 (Q9LI73) Chloroplast nucleoid DNA b... 40 0.001
>AV406732
Length = 422
Score = 149 bits (377), Expect = 8e-37
Identities = 72/140 (51%), Positives = 100/140 (71%), Gaps = 3/140 (2%)
Frame = +3
Query: 271 MCFGRDGIGRISFGDKGSLDQDETPFNVNPSHPTYNITINQVRVGTTLIDVEFTALFDSG 330
MCFG DG GRI+FGD GSLDQ +TPFN+ HPTYNITI Q++VG + D+EF A+FDSG
Sbjct: 3 MCFGSDGSGRITFGDTGSLDQGKTPFNLRALHPTYNITITQIKVGRNVADLEFHAIFDSG 182
Query: 331 TSFTYLVDPTYSRLSESFHSQVEDRR---RPPDSRIPFDYCYDMSPDSNTSLIPSMSLTM 387
TSFTYL DP Y+ +++ F+S ++ R + DS +PF++CY +SPD L P ++LTM
Sbjct: 183 TSFTYLNDPAYTLIADKFNSLIKADRPSSQSTDSDLPFEHCYAVSPDQTIEL-PYLNLTM 359
Query: 388 GGGSRFVVYDPIIIISTQSE 407
GG + V +P+I + ++ E
Sbjct: 360 KGGDDYYVTNPLIPVFSEVE 419
>TC14693 similar to GB|BAB01116.1|11994113|AB026658 CND41, chloroplast
nucleoid DNA binding protein-like {Arabidopsis
thaliana;} , partial (58%)
Length = 2067
Score = 95.1 bits (235), Expect = 2e-20
Identities = 92/366 (25%), Positives = 143/366 (38%), Gaps = 17/366 (4%)
Frame = +1
Query: 101 HYTTIELGTPGVKFMVALDTGSDLFWVPC-DCTRCSATRSSAFASALASDFDLSVYNPNG 159
+++ + +G P F + LDTGSD+ W+ C C C +++P
Sbjct: 511 YFSRVGVGQPAKPFYMVLDTGSDVNWLQCKPCADCYQQSD-------------PIFDPTT 651
Query: 160 SSTSKKVTCNNSLCTHRNQCLGTFSNCPYMVSYVSAETSTSGILVEDVLHLTQPDDNHDL 219
SST +TC+ C C Y VSY + + E + N
Sbjct: 652 SSTYGPLTCDAQQCQALEVSACRNGKCLYQVSYGDGSFTVGEFVTETMSFGGSGSVN--- 822
Query: 220 VEANVIFGCGQVQSGSFLDVAAPNGLFGLGMEKISVPSMLSREGFTADSFSMCFGRDGIG 279
V GCG G F+ A GL GLG +S+ S + A SFS C G
Sbjct: 823 ---RVAIGCGHDNEGLFVGAA---GLLGLGGGPLSLTSQIK-----ASSFSYCLVDRDSG 969
Query: 280 R---ISFGDKGSLDQDETPFNVNPSHPT-YNITINQVRVGTTLIDVEFTA---------- 325
+ + F D P N T Y + + V VG ++ +
Sbjct: 970 KSSTLEFNSARPGDSVTAPLLKNQKIGTFYYVELTGVSVGGQMVGIPPETFEMDQTGAGG 1149
Query: 326 -LFDSGTSFTYLVDPTYSRLSESFHSQVEDRRRPPDSRIPFDYCYDMSPDSNTSLIPSMS 384
+ DSGT+ T L Y+ + ++F ++ R + FD CYD+S + +P++S
Sbjct: 1150IIVDSGTAITRLRTQAYNSVRDAFRRLTQNLRSAEGVAL-FDTCYDLSTLQSVR-VPTVS 1323
Query: 385 LTMGGGSRFVVYDPIIIISTQSELVYCLAVV-KSAELNIIGQNFMTGYRVVFDRGKLILG 443
L GG + + +I S +C A ++ L+IIG G RV FD K +G
Sbjct: 1324LHFSGGKTWSLPAKNYLIPVDSAGTFCFAFAPTTSSLSIIGNVQQQGTRVSFDLAKNEVG 1503
Query: 444 WKKSDC 449
+ + C
Sbjct: 1504FSPNKC 1521
>TC8516 similar to UP|AAP72988 (AAP72988) CDR1, partial (27%)
Length = 1603
Score = 90.9 bits (224), Expect = 4e-19
Identities = 91/358 (25%), Positives = 142/358 (39%), Gaps = 29/358 (8%)
Frame = +1
Query: 105 IELGTPGVKFMVALDTGSDLFWV---PCDCTRCSATRSSAFASALASDFDLSVYNPNGSS 161
I +GTP V+ + DTGSDL WV PCD T+C A + +Y+P SS
Sbjct: 361 IYIGTPSVERLAIADTGSDLTWVQCSPCDNTKCFAQNT-------------PLYDPLNSS 501
Query: 162 TSKKVTCNNSLCT---HRNQCLGTFSNCPYMVSYVSAETSTSGILVEDVLHLTQPDDNHD 218
T + C++ CT + + +C Y +Y S G+ + + D
Sbjct: 502 TFTLLPCDSQPCTQLPYSQYVCSDYGDCIYAYTYGDNSYSYGGLSSDSI--------RFD 657
Query: 219 LVEA-----NVIFGCGQVQSGSFLDVAAPNGLFGLGMEKISVPSMLSREGFTADSFSMC- 272
A +FGCG + G+ GLG +S+ S L E FS C
Sbjct: 658 ATAATPTIPKFVFGCGFQNKFTADKSGKTTGIVGLGAGPLSLVSQLGDE--IGHKFSYCL 831
Query: 273 --FGRDGIGRISFGDKGSLDQD---ETPFNVNPSHPTYNITINQVRVGTTLI---DVEFT 324
F + ++ FG+ + + TP + P P Y + + + VG + +
Sbjct: 832 LPFSSNSNSKLKFGEAAIVQGNGVVSTPLIIKPDLPFYYLNLEGITVGAKTVKTGQTDGN 1011
Query: 325 ALFDSGTSFTYLVDPTYSRLSESFHSQV---EDRRRPPDSRIPFDYCYDMSPDSNTSLIP 381
+ DSG++ TYL + Y+ V ED+ P PFD+C+ +T P
Sbjct: 1012IIIDSGSTLTYLEESFYNEFVSLVKETVAVEEDQYIP----YPFDFCFTYKEGMSTP--P 1173
Query: 382 SMSLTMGGGSRFVVYDPIIIISTQSELVYCLAVVKS-----AELNIIGQ-NFMTGYRV 433
+ GG VV P+ + + + C VV S A +GQ +F GY +
Sbjct: 1174DVVFHFTGGD--VVLKPMNTLVLIEDNLICSTVVPSHFDGIAIFGNLGQIDFHVGYDI 1341
>TC8136 similar to GB|AAB60729.1|2160166|F21M12 F21M12.13 {Arabidopsis
thaliana;}, partial (70%)
Length = 1659
Score = 89.7 bits (221), Expect = 1e-18
Identities = 109/383 (28%), Positives = 158/383 (40%), Gaps = 27/383 (7%)
Frame = +1
Query: 81 AGLAFSDGNSTFRISSLGFLHYTTIELGTPGVKFMVALDTGSDLFWVPCD-CTRCSATRS 139
+G AF+ GN R+ ++GTPG + LDT +D +VPC CT CS T +
Sbjct: 331 SGQAFNIGNYIVRV-----------KIGTPGQLLFMVLDTSTDEAFVPCSGCTGCSTTTA 477
Query: 140 SAFASALASDFDLSVYNPNGSSTSKKVTCNNSLC--THRNQCLGTFS-NCPYMVSYVSAE 196
++P S+T + C+ LC C T S C + SY A
Sbjct: 478 P--------------FSPKASTTYSPLDCSVPLCGQVRGLSCPATGSATCSFNQSY--AG 609
Query: 197 TSTSGILVEDVLHLTQPDDNHDLVEANVIFGCGQVQSGSFLDVAAPNGLFGLGMEKISVP 256
++ S LV+D L L D V N FGC SG+ GL GLG +
Sbjct: 610 STFSATLVQDSLSLAT-----DAV-PNYSFGCINAISGA---TVPAQGLLGLGRGPL--- 753
Query: 257 SMLSREGFT-ADSFSMCF----GRDGIGRISFGDKGSLDQ-DETPFNVNPSHPT-YNITI 309
S+LS+ G + FS C G + G G TP NP P+ Y + +
Sbjct: 754 SLLSQTGTNYSGVFSYCLPSFKSYYFSGSLKLGPVGQPKSIRTTPLLRNPHRPSLYYVNL 933
Query: 310 NQVRVGTTLIDVEFTAL-----------FDSGTSFTYLVDPTYSRLSESFHSQVEDRRRP 358
+ VG L+ V +L DSGT T ++P Y+ + E F QV P
Sbjct: 934 TGISVGRVLVPVPAESLAFNPSTGAGTVIDSGTVITRFIEPVYAAVREEFRKQVTG---P 1104
Query: 359 PDSRIPFDYCYDMSPDSNTSLIPSMSLTMGGGSRFVVYDPIIIISTQSELVYCLAVVKSA 418
S FD C+ + + +L P ++L + G + + +I S+ L CLA+ +
Sbjct: 1105FSSLGAFDTCFVKTYE---TLAPVVTLHLEGLDLKLPLENSLIHSSSGSLA-CLAMAAAP 1272
Query: 419 E-----LNIIGQNFMTGYRVVFD 436
E LN+I RV+FD
Sbjct: 1273ENVNSVLNVIANYQQQNLRVLFD 1341
>BP057961
Length = 398
Score = 76.3 bits (186), Expect = 1e-14
Identities = 47/140 (33%), Positives = 68/140 (48%), Gaps = 6/140 (4%)
Frame = +3
Query: 31 HHRYSE----PVKKWSHSAPSPSHRWPEKGSVEYYAELADRD--RFLRGRRLSQFDAGLA 84
HHR+S + + H P R +YY + F+ R A L
Sbjct: 3 HHRFSAIRSXGIXGYXHELRLPXTR----ELXQYYXCFGPXEIVSFVARRLAGDQHAPLT 170
Query: 85 FSDGNSTFRISSLGFLHYTTIELGTPGVKFMVALDTGSDLFWVPCDCTRCSATRSSAFAS 144
F+ GN T ++ G+LH+ + +GTP + F+VALDTGSDLFW+PC+CT C S
Sbjct: 171 FAAGNDTHXDAAXGYLHFANVSVGTPPLWFVVALDTGSDLFWLPCNCTSCVRGLKRESGS 350
Query: 145 ALASDFDLSVYNPNGSSTSK 164
+ +L++Y SST K
Sbjct: 351 VI----NLNIYELEKSSTMK 398
>TC18309 similar to UP|Q9FG60 (Q9FG60) Aspartyl protease-like, partial (14%)
Length = 624
Score = 75.5 bits (184), Expect = 2e-14
Identities = 54/154 (35%), Positives = 80/154 (51%), Gaps = 5/154 (3%)
Frame = +2
Query: 68 DRFLRGRRLSQFDAGLAFSDGNSTFRISSLGFLHYTTIELGTPGVKFMVALDTGSDLFWV 127
D RGR LS D L GN ++ L ++T I LG+P + V +DTGSD+ WV
Sbjct: 173 DSSRRGRILSAVDFNLG---GNGLPTVTGL---YFTKIGLGSPSKDYYVQVDTGSDILWV 334
Query: 128 PC-DCTRCSATRSSAFASALASDFDLSVYNPNGSSTSKKVTCNNSLC--THRNQCLGTFS 184
C +CTRC R S L++Y+P S TS+ V+C ++ C T+ + LG +
Sbjct: 335 NCVECTRC--PRKSDIG------IGLTLYDPKRSKTSEFVSCEHNFCSSTYEGRILGCKA 490
Query: 185 N--CPYMVSYVSAETSTSGILVEDVLHLTQPDDN 216
CPY +SY ++T+G V+D L + + N
Sbjct: 491 ENPCPYSISYGDG-SATTGYYVQDYLTFNRVNGN 589
>TC16317 similar to UP|Q8W4C5 (Q8W4C5) Nucellin-like protein, partial (13%)
Length = 1131
Score = 72.0 bits (175), Expect = 2e-13
Identities = 64/258 (24%), Positives = 111/258 (42%), Gaps = 24/258 (9%)
Frame = +2
Query: 216 NHDLVEANVIFGCGQVQSGSFLD-VAAPNGLFGLGMEKISVPSMLSREGFTADSFSMCFG 274
N + N +FGCG Q+GS L+ ++ +G+ GL K+S+P L+ +G + C
Sbjct: 23 NGSKTKFNFVFGCGYDQAGSLLNTLSKTDGIMGLSRAKVSLPYQLASKGLIKNVVGHCLS 202
Query: 275 RDGIGR--ISFGDKGSLDQDET--PFNVNPSHPTYNITINQVRVGTTLIDVEFTA----- 325
D +G + GD + T P S Y I + G + + +
Sbjct: 203 SDAVGGGYMFLGDDFVPNWGMTWVPMAYTHSSDLYQTEILGINYGNHPLSSDGHSKVRKV 382
Query: 326 LFDSGTSFTYLVDPTYSRLSESFHSQVEDRRRPPDSRIPFDYCY-DMSPDSNTSLIPSM- 383
+FDSG+S+TY Y L S R DS F C+ D P + +
Sbjct: 383 VFDSGSSYTYFPKEAYLDLVASLKEVSGLRLIQDDSDTTFPICWQDDFPIRSVKDVKDFF 562
Query: 384 -SLTMGGGSRFVVYDPIIIISTQSELVY------CLAVVKSAELN-----IIGQNFMTGY 431
+LT+ G+++ + I + L+ CLA++ + ++ I+G + G+
Sbjct: 563 KTLTLRFGNKWWILSTFFQIPPEGYLIISKKGNVCLAILDGSNVHDGSSIILGDVSLRGH 742
Query: 432 RVVFDRGKLILGWKKSDC 449
VV+D +GW+++DC
Sbjct: 743 LVVYDNVNQRIGWERADC 796
>TC15942 similar to UP|Q9FL43 (Q9FL43) Nucleoid DNA-binding-like protein,
partial (66%)
Length = 973
Score = 67.8 bits (164), Expect = 4e-12
Identities = 70/255 (27%), Positives = 105/255 (40%), Gaps = 19/255 (7%)
Frame = +1
Query: 106 ELGTPGVKFMVALDTGSDLFWVPCD-CTRCSATRSSAFASALASDFDLSVYNPNGSSTSK 164
++G P ++A+DT +D WVPC C CS+ +++ P S+T K
Sbjct: 298 KIGNPPQTLLMAMDTSNDAAWVPCSGCDGCSS----------------ALFAPEKSTTFK 429
Query: 165 KVTCNNSLCTHRNQCLGTFSNCPYMVSYVSAETSTSGILVEDVLHL-TQPDDNHDLVEAN 223
V C + C S C + +Y S +S + L +D + L T P ++
Sbjct: 430 SVGCAAAECKQVPNPSCGVSACNFNFTYGS--SSIAANLAQDTVTLATDPVPSY------ 585
Query: 224 VIFGCGQVQSGSFLDVAAPNGLFGLGMEKISVPSMLSREGFTADSFSMCF----GRDGIG 279
FGC +GS P GL GLG +S+ S + +FS C + G
Sbjct: 586 -TFGCVAKTTGS---SNPPQGLLGLGRGPLSLLS--QTQNLYKSTFSYCLPSFKSLNFSG 747
Query: 280 RISFGDKGS-LDQDETPFNVNPSHPT-YNITINQVRVGTTLIDVEFTAL----------- 326
+ G L TP NP P+ Y + + +RVG ++D+ AL
Sbjct: 748 SLRLGPVAQPLRIKFTPLLKNPRRPSLYYVNLVAIRVGRKIVDIPPAALAFNPTTGAGTI 927
Query: 327 FDSGTSFTYLVDPTY 341
FDSGT +T L P Y
Sbjct: 928 FDSGTVYTRLAAPAY 972
>TC10379 weakly similar to UP|Q8H9F4 (Q8H9F4) 41 kDa chloroplast nucleoid
DNA binding protein (CND41), partial (43%)
Length = 1002
Score = 67.0 bits (162), Expect = 7e-12
Identities = 53/192 (27%), Positives = 76/192 (38%), Gaps = 8/192 (4%)
Frame = +3
Query: 101 HYTTIELGTPGVKFMVALDTGSDLFWVPCD-CTRCSATRSSAFASALASDFDLSVYNPNG 159
++ + LGTP + DTGSDL W C C R + +++P
Sbjct: 429 YFVVVGLGTPKSDLSLVFDTGSDLTWTQCQPCARSCYKQQD------------PIFDPKR 572
Query: 160 SSTSKKVTCNNSLCTHRNQ-------CLGTFSNCPYMVSYVSAETSTSGILVEDVLHLTQ 212
S++ +TC +S CT C C Y + Y + S G + L +T
Sbjct: 573 STSYYNITCTSSDCTQLTSATGNDPACATITKACVYGIQYGDSSFSV-GFFSRERLTVTP 749
Query: 213 PDDNHDLVEANVIFGCGQVQSGSFLDVAAPNGLFGLGMEKISVPSMLSREGFTADSFSMC 272
D V N +FGCGQ G F A GL GLG IS S++ S+ +
Sbjct: 750 TD-----VVDNFLFGCGQNNQGLFNGAA---GLLGLGRHPISFVQQTSQKYQKKFSYCLP 905
Query: 273 FGRDGIGRISFG 284
+G +SFG
Sbjct: 906 STTSAVGHLSFG 941
>TC17357 similar to UP|Q9FFC3 (Q9FFC3) Protease-like protein, partial (24%)
Length = 577
Score = 64.3 bits (155), Expect = 4e-11
Identities = 48/145 (33%), Positives = 68/145 (46%), Gaps = 8/145 (5%)
Frame = +1
Query: 52 WPEKGSVEYYAELADRDRFLRGRR--LSQFDAGLAFSDGNSTFRISSLGFLHYTTIELGT 109
+P VE ++L RD LR RR L + G+A TF +G L++T +++GT
Sbjct: 175 FPTNHGVEL-SQLRARD-MLRHRRMLLQSSNNGVADFSVQGTFDPFQVG-LYFTRVQIGT 345
Query: 110 PGVKFMVALDTGSDLFWVPC-DCTRCSATRSSAFASALASDFDLSVYNPNGSSTSKKVTC 168
P V+F V +DTGSD+ WV C C C T L ++P SSTS ++C
Sbjct: 346 PPVEFYVQIDTGSDVLWVSCSSCNGCPQTS--------GLQIQLDFFDPGHSSTSSLISC 501
Query: 169 NNSLC-----THRNQCLGTFSNCPY 188
++ C T C G C Y
Sbjct: 502 SDQRCNGGIQTSDASCSGRDGQCSY 576
>TC18588
Length = 390
Score = 60.8 bits (146), Expect = 5e-10
Identities = 37/97 (38%), Positives = 57/97 (58%), Gaps = 3/97 (3%)
Frame = +3
Query: 1 MLSFTKIIVIILIILHL-SMCCNAH-IFTFTMHHRYSEPVKKWSHSAPSPSHRWPEKGSV 58
+++ +++++ L+++ L S CC+ F F +HHR+S+PVK R P KG+
Sbjct: 108 LMALPQLVLLGLVVVSLASQCCHGSGQFGFDIHHRFSDPVKG---ILGIDELRLPHKGTP 278
Query: 59 EYYAELADRDRFLRGRRLS-QFDAGLAFSDGNSTFRI 94
+YYA L RDR RGRRL+ A L F+ GN T +I
Sbjct: 279 QYYAALVHRDRVFRGRRLAGDQHAPLTFAAGNDTHQI 389
>TC14194 weakly similar to UP|Q8GT67 (Q8GT67) Xyloglucan-specific fungal
endoglucanase inhibitor protein precursor, partial (11%)
Length = 892
Score = 57.8 bits (138), Expect = 4e-09
Identities = 63/274 (22%), Positives = 104/274 (36%), Gaps = 25/274 (9%)
Frame = +3
Query: 101 HYTTIELGTPGVKFMVALDTGSDLFWVPCDCTRCSATRSSAFASALASDFDLSVYNPNGS 160
+YT+I +GTP K +A+D + W CD TR YN S
Sbjct: 159 YYTSIGIGTPNHKLNLAIDLAGEFLWYDCD-TR---------------------YN---S 263
Query: 161 STSKKVTCNNSLCTHRNQCLG----------TFSNCPYMVSYVSAETSTSGILVEDVLHL 210
S+ V C+ C + C+G T + C ++ A+T SG + ED+LH+
Sbjct: 264 SSYLPVPCDTQKCPQNSPCIGCNGFPTKPGCTNNTCGLSITNPFADTIFSGDMGEDLLHI 443
Query: 211 TQPDDNHDLVEANVIFGCGQVQSGSFLD------VAAPNGLFGLGMEKISVPSMLSREGF 264
Q ++ F G S F G+ GL ++S+P+ +S
Sbjct: 444 PQ-------IKVPRSFASGCADSDRFSTPLLVGLAKGTKGILGLARSQLSLPTQISSSYN 602
Query: 265 TADSFSMCF---GRDGIGRISFGDKGSLDQDETPFN-VNPSHPTYNITINQVRVGTTLID 320
F++C G G+I G + S + S Y I + + V +++
Sbjct: 603 VPPKFTLCLPSSNTKGTGKIFIGGRPSSRANVARIGFALTSSEEYFINVKSIMVDDKVVN 782
Query: 321 VEFTALF-----DSGTSFTYLVDPTYSRLSESFH 349
+ + L + GT + L P Y+ L S +
Sbjct: 783 FDTSLLSLDKNGNGGTKISTLGTP-YTVLHNSIY 881
>TC8779 similar to UP|Q8L934 (Q8L934) Nucleoid DNA-binding-like protein,
partial (27%)
Length = 737
Score = 52.4 bits (124), Expect = 2e-07
Identities = 41/134 (30%), Positives = 64/134 (47%), Gaps = 5/134 (3%)
Frame = +3
Query: 327 FDSGTSFTYLVDPTYSRLSESFHSQVEDRRRPPDSRIPFDYCYDMSPDSNTSLIPSMSLT 386
FDSGT FT LV+P Y+ + F +V RR P + FD CY + + ++P ++
Sbjct: 3 FDSGTVFTRLVEPAYTAVRNEFRRRV-GRRVPVTTLGGFDTCY-----TGSIVVPPITFM 164
Query: 387 MGGGSRFVVYDPIIIISTQSELVYCLAVV-----KSAELNIIGQNFMTGYRVVFDRGKLI 441
G + + D I+I ST + CLA+ ++ LN+I +RV++D
Sbjct: 165 FSGMNVTLPTDNIVIHSTAGSIT-CLAMAVTPNNVNSVLNVIANMQQQNHRVLYDVPNSR 341
Query: 442 LGWKKSDCKWLFFC 455
LG + C L C
Sbjct: 342 LGIARELCS*L*SC 383
>TC14947 similar to GB|AAP31963.1|30387595|BT006619 At1g01300 {Arabidopsis
thaliana;}, partial (26%)
Length = 728
Score = 50.1 bits (118), Expect = 8e-07
Identities = 36/125 (28%), Positives = 58/125 (45%), Gaps = 1/125 (0%)
Frame = +2
Query: 326 LFDSGTSFTYLVDPTYSRLSESFHSQVEDRRRPPDSRIPFDYCYDMSPDSNTSLIPSMSL 385
+ DSGTS T L P Y L ++F +R P+ + FD C+D+S + +P++ L
Sbjct: 17 IIDSGTSVTRLTRPGYIALRDAFRLGALHLKRAPEFSL-FDTCFDLSGQTEVK-VPTVVL 190
Query: 386 TMGGGS-RFVVYDPIIIISTQSELVYCLAVVKSAELNIIGQNFMTGYRVVFDRGKLILGW 444
G + +I + + + A S L+IIG G+RVV+D G +G+
Sbjct: 191 HFRGADVSLPATNYLIPVDSSGSFCFAFAGTMSG-LSIIGNIQQQGFRVVYDLGSSRVGF 367
Query: 445 KKSDC 449
C
Sbjct: 368 APRGC 382
>AV409850
Length = 419
Score = 47.0 bits (110), Expect = 7e-06
Identities = 32/76 (42%), Positives = 43/76 (56%)
Frame = +1
Query: 52 WPEKGSVEYYAELADRDRFLRGRRLSQFDAGLAFSDGNSTFRISSLGFLHYTTIELGTPG 111
+P VE +EL RD LR RR Q + + TF S +G L+YT ++LGTP
Sbjct: 199 FPSNHGVEL-SELRARD-MLRHRRFLQSVSYVVDFPVKGTFDPSQVG-LYYTKVKLGTPP 369
Query: 112 VKFMVALDTGSDLFWV 127
+F V +DTGSD+ WV
Sbjct: 370 REFYVQIDTGSDVLWV 417
>TC15949 similar to UP|Q9LTW4 (Q9LTW4) Chloroplast nucleoid DNA binding
protein-like, partial (6%)
Length = 690
Score = 46.6 bits (109), Expect = 9e-06
Identities = 41/161 (25%), Positives = 70/161 (43%), Gaps = 12/161 (7%)
Frame = +1
Query: 305 YNITINQVRVGTTLIDV-----EFTA----LFDSGTSFTYLVDPTYSRLSESFHSQVEDR 355
Y + + + VG ++ + +F A + DSGT+ T L P Y +L E+ +
Sbjct: 4 YGVNVVGISVGGQMLKIPSQVWDFNAQGGTIIDSGTTLTNLALPAYEQLFEALKKSLTKV 183
Query: 356 RRPPDSRI-PFDYCYDMSPDSNTSLIPSMSLTMGGGSRF--VVYDPIIIISTQSELVYCL 412
+R P DYC+D + + S +P + GG RF V II ++ Q + + L
Sbjct: 184 KRVPAGDFGGLDYCFD-AKGFDESSVPRLVFHFAGGVRFEPPVKSYIIDVAPQVKCIGVL 360
Query: 413 AVVKSAELNIIGQNFMTGYRVVFDRGKLILGWKKSDCKWLF 453
A + ++IG + FD +G+ S C +F
Sbjct: 361 A-INGPGASVIGNIMQQNHLWEFDLAHNTVGFAPSACN*IF 480
>TC15943 similar to UP|Q8GT67 (Q8GT67) Xyloglucan-specific fungal
endoglucanase inhibitor protein precursor, partial (15%)
Length = 968
Score = 44.7 bits (104), Expect = 4e-05
Identities = 57/264 (21%), Positives = 99/264 (36%), Gaps = 37/264 (14%)
Frame = +2
Query: 100 LHYTTIELGTPGVKFMVALDTGSDLFWVPCDCTRCSATRSSAFASALASDFDLSVYNPNG 159
L YT++ +GTP F + +D G + W C+ N
Sbjct: 191 LFYTSVGVGTPRHNFDLVIDLGGPILWYDCN-------------------------NHYN 295
Query: 160 SSTSKKVTCNNSLCTHR----NQCLG------TFSNCPYMVSYVSAETSTSGILVEDVLH 209
SST + V+C++ C++ C G T + C + A SG EDVL
Sbjct: 296 SSTYRPVSCDSKSCSNTGAGCTSCDGPLKPGCTNNTCGANILNPLANVIFSGDTGEDVLF 475
Query: 210 LTQPDDNHDLVEANVIFGCGQVQSGSFLD--------VAAPNGLFGLGMEKISVPSMLSR 261
++ +V+ GC S +F D G+ GL ++++P+ LS
Sbjct: 476 ISSS-------IVSVLSGC--ASSDAFNDDPPLLKGLPQTSKGILGLARTQLALPTQLSS 628
Query: 262 EGFTADSFSMCF---GRDGIGRISFGD-KGSLDQD----ETPFNVNP-----------SH 302
F++C ++G+G + G SL TP +NP +
Sbjct: 629 SEL-PHKFALCLPSSNKNGLGNLFLGGAPDSLPHQPALLTTPLIINPFSTAPIFSEGDAS 805
Query: 303 PTYNITINQVRVGTTLIDVEFTAL 326
Y I + +++G ++D + + L
Sbjct: 806 YEYFIVVKSIKIGGEVLDFKSSLL 877
>TC16802 similar to GB|AAP31963.1|30387595|BT006619 At1g01300 {Arabidopsis
thaliana;}, partial (28%)
Length = 812
Score = 44.3 bits (103), Expect = 5e-05
Identities = 37/140 (26%), Positives = 52/140 (36%), Gaps = 3/140 (2%)
Frame = +3
Query: 101 HYTTIELGTPGVKFMVALDTGSDLFWVPC-DCTRCSATRSSAFASALASDFDLSVYNPNG 159
++T I +GTP + LDTGSD+ W+ C C +C + V++P+
Sbjct: 450 YFTRIGVGTPARYVYMVLDTGSDVVWLQCAPCRKCYSQAD-------------PVFDPSK 590
Query: 160 SSTSKKVTCNNSLCTHRNQCLGTFSN--CPYMVSYVSAETSTSGILVEDVLHLTQPDDNH 217
S T + C LC R G S C Y VSY + E +
Sbjct: 591 SRTYAGIPCGAPLC-RRLDSAGCNSKKVCQYQVSYGDGSFTFGDFSTETLTFRRSR---- 755
Query: 218 DLVEANVIFGCGQVQSGSFL 237
V GCG G F+
Sbjct: 756 ---VTRVALGCGHDNEGLFV 806
>AV427004
Length = 428
Score = 41.6 bits (96), Expect = 3e-04
Identities = 30/90 (33%), Positives = 42/90 (46%), Gaps = 2/90 (2%)
Frame = +3
Query: 198 STSGILVEDVLHLTQPDDNHDLVEANVIFGCGQVQSGSFLDVAAPN--GLFGLGMEKISV 255
S+ G+LV D +HL N +V + FGCG Q S + P+ G+ GLG + S+
Sbjct: 27 SSLGVLVRDHIHLHFT--NGSVVRPKIAFGCGYDQKYSG-PITPPSTAGVIGLGNGRSSI 197
Query: 256 PSMLSREGFTADSFSMCFGRDGIGRISFGD 285
S L G + C G G + FGD
Sbjct: 198 VSQLHSLGLIRNVVGHCLSAQGGGFLFFGD 287
>TC10566 similar to UP|Q9LI73 (Q9LI73) Chloroplast nucleoid DNA binding
protein-like, nucellin-like protein, partial (25%)
Length = 774
Score = 39.7 bits (91), Expect = 0.001
Identities = 32/128 (25%), Positives = 60/128 (46%), Gaps = 6/128 (4%)
Frame = +2
Query: 328 DSGTSFTYLVDPTYSRLSESFHSQVEDRRRP--PDSRIPFDYCYDMSPDSNTSLIPSMSL 385
DSGT+ T+L +P Y ++ +F +V R P D + FD C ++ S + + L
Sbjct: 50 DSGTTLTFLAEPAYRQILAAFRRRV---RLPAVEDPSLAFDLCVNV---SGVARVKFPKL 211
Query: 386 TMGGGSRFVVYDPIIIISTQ-SELVYCLAVVKS---AELNIIGQNFMTGYRVVFDRGKLI 441
+G + V+ P + ++ V CLA+ + + ++IG GY F+ +
Sbjct: 212 RIGLAGKSVLSPPARNYFIEVADRVKCLAIQPAKPGSGFSVIGNLMQQGYLFQFEVDRSR 391
Query: 442 LGWKKSDC 449
+G+ + C
Sbjct: 392 VGFSRRGC 415
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.323 0.137 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,535,279
Number of Sequences: 28460
Number of extensions: 141244
Number of successful extensions: 982
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 956
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 963
length of query: 492
length of database: 4,897,600
effective HSP length: 94
effective length of query: 398
effective length of database: 2,222,360
effective search space: 884499280
effective search space used: 884499280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC149547.3


