

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149546.1 - phase: 0
(133 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
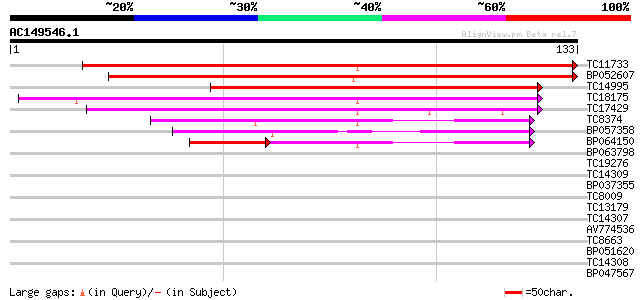
Score E
Sequences producing significant alignments: (bits) Value
TC11733 weakly similar to UP|Q93XF3 (Q93XF3) Kinesin heavy chain... 177 5e-46
BP052607 146 1e-36
TC14995 similar to UP|Q9SVJ8 (Q9SVJ8) Kinesin like protein, part... 94 8e-21
TC18175 similar to UP|Q8L5J2 (Q8L5J2) Kinesin-like protein, part... 88 6e-19
TC17429 similar to UP|Q8S950 (Q8S950) Kinesin-like protein NACK1... 82 3e-17
TC8374 45 4e-06
BP057358 42 4e-05
BP064150 37 6e-05
BP063798 32 0.048
TC19276 26 2.0
TC14309 homologue to UP|H2B_GOSHI (O22582) Histone H2B, partial ... 25 3.5
BP037355 25 4.5
TC8009 homologue to UP|Q9LKJ2 (Q9LKJ2) Cytosolic phosphoglycerat... 25 4.5
TC13179 similar to UP|Q9ZVA9 (Q9ZVA9) F9K20.4 protein, partial (5%) 25 5.9
TC14307 homologue to UP|H2B_GOSHI (O22582) Histone H2B, partial ... 25 5.9
AV774536 25 5.9
TC8663 homologue to UP|R10B_ARATH (P59230) 60S ribosomal protein... 24 7.7
BP051620 24 7.7
TC14308 homologue to UP|H2B_GOSHI (O22582) Histone H2B, partial ... 24 7.7
BP047567 24 7.7
>TC11733 weakly similar to UP|Q93XF3 (Q93XF3) Kinesin heavy chain
(Fragment), partial (11%)
Length = 628
Score = 177 bits (449), Expect = 5e-46
Identities = 84/117 (71%), Positives = 104/117 (88%), Gaps = 1/117 (0%)
Frame = +3
Query: 18 EDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWGISMSSKHRRLQLAHRLW 77
EDGRTLTPESS R L+RERQMLS+ MQ+KLSKSER N+YL+W + +S+KHR LQLAHRLW
Sbjct: 3 EDGRTLTPESSMRSLKRERQMLSKPMQKKLSKSERMNLYLRWVLRLSTKHRSLQLAHRLW 182
Query: 78 SET-DINHVRESATIVAKLVGTVEPDQAFKEMFGLNFAPRRRRKKSFGWTSSMKHIL 133
S+T D+ H+R+SA +VAKLVG+VEP+QAFKEMFGLNFAPR +KSFGWT+S++H+L
Sbjct: 183 SDTKDMEHIRDSAALVAKLVGSVEPEQAFKEMFGLNFAPRPSSRKSFGWTASVRHLL 353
>BP052607
Length = 466
Score = 146 bits (368), Expect = 1e-36
Identities = 71/111 (63%), Positives = 89/111 (79%), Gaps = 1/111 (0%)
Frame = -2
Query: 24 TPESSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWGISMSSKHRRLQLAHRLWSE-TDI 82
TPESS R L RE MLS+ M +KLSKSER N+YL+WG+ +S KHR L LAHRLWS+ D+
Sbjct: 465 TPESSLRSLXREXPMLSQPMPKKLSKSERMNLYLRWGLPLSPKHRSLPLAHRLWSDPKDL 286
Query: 83 NHVRESATIVAKLVGTVEPDQAFKEMFGLNFAPRRRRKKSFGWTSSMKHIL 133
H+R+SA VAKLVG+VEP+ AFKEMFGL+FAPR +KSFGWT+ ++H+L
Sbjct: 285 EHLRDSAARVAKLVGSVEPEPAFKEMFGLHFAPRPSSRKSFGWTARVRHLL 133
>TC14995 similar to UP|Q9SVJ8 (Q9SVJ8) Kinesin like protein, partial (4%)
Length = 783
Score = 94.0 bits (232), Expect = 8e-21
Identities = 41/78 (52%), Positives = 59/78 (75%)
Frame = +3
Query: 48 SKSERDNMYLKWGISMSSKHRRLQLAHRLWSETDINHVRESATIVAKLVGTVEPDQAFKE 107
S+ ER ++ +W I ++SK RR+QLA+RLWS TD+NH+ +SAT+VAKLV E +A KE
Sbjct: 3 SEEERKRLFKEWDIPLNSKRRRMQLANRLWSNTDMNHIMQSATVVAKLVRFSEQGKALKE 182
Query: 108 MFGLNFAPRRRRKKSFGW 125
MFGL+F P+ R++S+ W
Sbjct: 183 MFGLSFTPQLTRRRSYSW 236
>TC18175 similar to UP|Q8L5J2 (Q8L5J2) Kinesin-like protein, partial (15%)
Length = 608
Score = 87.8 bits (216), Expect = 6e-19
Identities = 50/133 (37%), Positives = 79/133 (58%), Gaps = 10/133 (7%)
Frame = +1
Query: 3 VELRRLSYLKDN---------QILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERD 53
VE+RRL++L+ + +L D + +S R L++ER+ LS+++ KL+ ER+
Sbjct: 1 VEIRRLTWLEQHLAELGNASPALLGDEPAGSVSASIRALKQEREYLSKRVNTKLTAEERE 180
Query: 54 NMYLKWGISMSSKHRRLQLAHRLWSET-DINHVRESATIVAKLVGTVEPDQAFKEMFGLN 112
+Y KW + K RRLQL ++ W++ ++ HV+ESA IVAKL+ D+ K+MF LN
Sbjct: 181 YLYAKWEVPPVGKQRRLQLVNKFWTDPYNLQHVQESAEIVAKLIDFCVSDENSKDMFELN 360
Query: 113 FAPRRRRKKSFGW 125
FA +K GW
Sbjct: 361 FASPYNKKTWGGW 399
>TC17429 similar to UP|Q8S950 (Q8S950) Kinesin-like protein NACK1, partial
(14%)
Length = 587
Score = 82.0 bits (201), Expect = 3e-17
Identities = 46/110 (41%), Positives = 66/110 (59%), Gaps = 3/110 (2%)
Frame = +1
Query: 19 DGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWGISMSSKHRRLQLAHRLWS 78
D T++ SS R L+RER+ L+++M + + ER+ +Y KW + + K R+LQ +LW+
Sbjct: 70 DEPTISLSSSLRALKREREFLAKRMTSRFTLEEREELYYKWDVPLEGKQRKLQFVSKLWT 249
Query: 79 ET-DINHVRESATIVAKLVG-TVEPDQAFKEMFGLNFA-PRRRRKKSFGW 125
+ D HV+ESA IVAKLVG + KEMF LNF P +R GW
Sbjct: 250 DPHDQKHVQESAEIVAKLVGLSSTRGNMSKEMFELNFVLPSDKRPWLMGW 399
>TC8374
Length = 601
Score = 45.1 bits (105), Expect = 4e-06
Identities = 30/92 (32%), Positives = 47/92 (50%), Gaps = 2/92 (2%)
Frame = +2
Query: 34 RERQMLSRQMQRKLSKSERDNMY-LKWGISMSSKHRRLQLAHRLWSET-DINHVRESATI 91
+ERQ+LS + ++ + ++ + + +K QLAHRLW++ D+ H +S T
Sbjct: 191 KERQILSTHAEEEIKIW*NEPIFEMVPSFELEAKEFTTQLAHRLWTDIEDMEHFTKSGT- 367
Query: 92 VAKLVGTVEPDQAFKEMFGLNFAPRRRRKKSF 123
FKE+FGLNFAP +KSF
Sbjct: 368 -------------FKEIFGLNFAPPFSSRKSF 424
>BP057358
Length = 473
Score = 42.0 bits (97), Expect = 4e-05
Identities = 29/87 (33%), Positives = 42/87 (47%), Gaps = 2/87 (2%)
Frame = -1
Query: 39 LSRQMQRKLSKSERDNMYLKWG--ISMSSKHRRLQLAHRLWSETDINHVRESATIVAKLV 96
+ + ++ SK R N Y + G + + +K QLAHR W TDI +
Sbjct: 377 IDAHLPKRKSKFGRMNPYFEMGPSLELEAKEFTTQLAHRPW--TDIEDL----------- 237
Query: 97 GTVEPDQAFKEMFGLNFAPRRRRKKSF 123
G + FKE+FGL+FAP +KSF
Sbjct: 236 GPLTKSGTFKEIFGLHFAPPFSSRKSF 156
>BP064150
Length = 422
Score = 36.6 bits (83), Expect(2) = 6e-05
Identities = 23/64 (35%), Positives = 32/64 (49%), Gaps = 1/64 (1%)
Frame = -3
Query: 61 ISMSSKHRRLQLAHRLWSET-DINHVRESATIVAKLVGTVEPDQAFKEMFGLNFAPRRRR 119
+ + K QLAHRLW++ D+ +S T FKE+FGL+FAP
Sbjct: 327 LELEVKEFTTQLAHRLWTDIEDLEPFTKSGT--------------FKEIFGLHFAPPFSS 190
Query: 120 KKSF 123
+KSF
Sbjct: 189 RKSF 178
Score = 23.9 bits (50), Expect(2) = 6e-05
Identities = 8/19 (42%), Positives = 14/19 (73%)
Frame = -2
Query: 43 MQRKLSKSERDNMYLKWGI 61
+Q++ SKS R N+Y +W +
Sbjct: 385 LQKRKSKSGRMNLYSRWAL 329
>BP063798
Length = 289
Score = 31.6 bits (70), Expect = 0.048
Identities = 15/30 (50%), Positives = 22/30 (73%), Gaps = 1/30 (3%)
Frame = -2
Query: 68 RRLQLAHRLWSET-DINHVRESATIVAKLV 96
RRL L ++ W++ ++ HV ESA IVAKL+
Sbjct: 288 RRLPLVNKFWTDPYNLQHVPESAEIVAKLI 199
>TC19276
Length = 494
Score = 26.2 bits (56), Expect = 2.0
Identities = 17/64 (26%), Positives = 30/64 (46%)
Frame = +2
Query: 16 ILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWGISMSSKHRRLQLAHR 75
+L+DG E + +R Q+ +Q+ + E + LKW S K R ++ H+
Sbjct: 95 VLQDGGKSAWEKKQWMKKRMVQLDEQQVSYRAEAFELEKPRLKWARFSSQKEREME-RHQ 271
Query: 76 LWSE 79
L +E
Sbjct: 272 LENE 283
>TC14309 homologue to UP|H2B_GOSHI (O22582) Histone H2B, partial (93%)
Length = 682
Score = 25.4 bits (54), Expect = 3.5
Identities = 16/63 (25%), Positives = 32/63 (50%)
Frame = +3
Query: 26 ESSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWGISMSSKHRRLQLAHRLWSETDINHV 85
E+SK +R R+ + Q + + + +++L+ + S +HR LQ H + H+
Sbjct: 177 EASKGGRKRRRRQEEEEEQEE--RGDIQDLHLQGAEAGSPRHRYLQQGHGDHEQFHQRHL 350
Query: 86 RES 88
RE+
Sbjct: 351 REA 359
>BP037355
Length = 422
Score = 25.0 bits (53), Expect = 4.5
Identities = 10/39 (25%), Positives = 16/39 (40%)
Frame = -2
Query: 54 NMYLKWGISMSSKHRRLQLAHRLWSETDINHVRESATIV 92
N Y KW +R+ Q+A +W + S +V
Sbjct: 421 NQYRKWNYKSKITNRKSQMARAIWLSMCVTGAVSSGQVV 305
>TC8009 homologue to UP|Q9LKJ2 (Q9LKJ2) Cytosolic phosphoglycerate kinase
, complete
Length = 1697
Score = 25.0 bits (53), Expect = 4.5
Identities = 10/19 (52%), Positives = 13/19 (67%)
Frame = +3
Query: 64 SSKHRRLQLAHRLWSETDI 82
S+K R LQL H LW +T +
Sbjct: 867 STKPRVLQLGHHLWRKTSL 923
>TC13179 similar to UP|Q9ZVA9 (Q9ZVA9) F9K20.4 protein, partial (5%)
Length = 533
Score = 24.6 bits (52), Expect = 5.9
Identities = 11/35 (31%), Positives = 19/35 (53%)
Frame = +2
Query: 60 GISMSSKHRRLQLAHRLWSETDINHVRESATIVAK 94
G + ++ + AHR WS D++ V ES+ + K
Sbjct: 56 GTPIVGDYKYVWQAHRKWSHFDLSDVEESSEELLK 160
>TC14307 homologue to UP|H2B_GOSHI (O22582) Histone H2B, partial (90%)
Length = 701
Score = 24.6 bits (52), Expect = 5.9
Identities = 18/79 (22%), Positives = 34/79 (42%), Gaps = 7/79 (8%)
Frame = +1
Query: 17 LEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERD-------NMYLKWGISMSSKHRR 69
++ GR +P + R E R R+ + E+ +++L+ + S +HR
Sbjct: 121 VDGGREGSPGGEETQGREEASQGGRSRCRRQEEEEKQEERGDLQDLHLQGAEAGSPRHRY 300
Query: 70 LQLAHRLWSETDINHVRES 88
LQ H + H+RE+
Sbjct: 301 LQQGHGDHEQFHQRHLREA 357
>AV774536
Length = 451
Score = 24.6 bits (52), Expect = 5.9
Identities = 10/23 (43%), Positives = 14/23 (60%)
Frame = -1
Query: 111 LNFAPRRRRKKSFGWTSSMKHIL 133
+ F P R + SFG TS + H+L
Sbjct: 268 IRFLPERWKVSSFGTTSPLFHVL 200
>TC8663 homologue to UP|R10B_ARATH (P59230) 60S ribosomal protein L10a-2,
partial (97%)
Length = 993
Score = 24.3 bits (51), Expect = 7.7
Identities = 17/56 (30%), Positives = 26/56 (46%)
Frame = +3
Query: 72 LAHRLWSETDINHVRESATIVAKLVGTVEPDQAFKEMFGLNFAPRRRRKKSFGWTS 127
L RL S T I + ++ + +++P F + LN + RRRR GW S
Sbjct: 165 LNSRLVSRTMIPRKTSVSVALSSCLISLDPR*RFACLVMLNTSKRRRRLDWTGWMS 332
>BP051620
Length = 545
Score = 24.3 bits (51), Expect = 7.7
Identities = 8/19 (42%), Positives = 12/19 (63%)
Frame = +3
Query: 115 PRRRRKKSFGWTSSMKHIL 133
PR R FGWT S++ ++
Sbjct: 222 PRXRAGSHFGWTQSVRQLI 278
>TC14308 homologue to UP|H2B_GOSHI (O22582) Histone H2B, partial (96%)
Length = 764
Score = 24.3 bits (51), Expect = 7.7
Identities = 15/55 (27%), Positives = 27/55 (48%), Gaps = 2/55 (3%)
Frame = +2
Query: 36 RQMLSRQMQRKLSKSERD--NMYLKWGISMSSKHRRLQLAHRLWSETDINHVRES 88
R RQ + + + RD +++L+ + S +HR LQ H + H+RE+
Sbjct: 203 RSRCRRQEEEEKQEERRDLQDLHLQGPEAGSPRHRYLQQGHGDHEQLHQRHLREA 367
>BP047567
Length = 480
Score = 24.3 bits (51), Expect = 7.7
Identities = 9/16 (56%), Positives = 12/16 (74%)
Frame = +2
Query: 114 APRRRRKKSFGWTSSM 129
+PRRRR + F W SS+
Sbjct: 173 SPRRRRPRLFPWLSSL 220
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.131 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,992,244
Number of Sequences: 28460
Number of extensions: 19769
Number of successful extensions: 141
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 134
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 134
length of query: 133
length of database: 4,897,600
effective HSP length: 80
effective length of query: 53
effective length of database: 2,620,800
effective search space: 138902400
effective search space used: 138902400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 50 (23.9 bits)
Medicago: description of AC149546.1


