

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149490.2 - phase: 1
(317 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
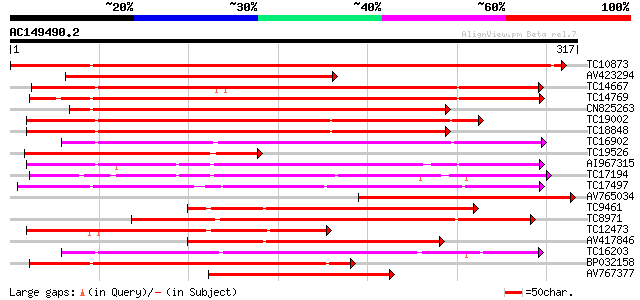
Score E
Sequences producing significant alignments: (bits) Value
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 386 e-108
AV423294 278 9e-76
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 241 1e-64
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 225 7e-60
CN825263 224 2e-59
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 220 3e-58
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 209 4e-55
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 204 1e-53
TC19526 homologue to UP|AAQ96340 (AAQ96340) Protein kinase-like ... 197 2e-51
AI967315 192 6e-50
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 191 1e-49
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 190 2e-49
AV765034 189 4e-49
TC9461 similar to UP|O49840 (O49840) Protein kinase (AT2G02800/T... 183 3e-47
TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P... 178 1e-45
TC12473 similar to UP|Q9M068 (Q9M068) Protein kinase-like protei... 176 4e-45
AV417846 169 6e-43
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 167 2e-42
BP032158 166 5e-42
AV767377 165 8e-42
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 386 bits (992), Expect = e-108
Identities = 186/311 (59%), Positives = 244/311 (77%)
Frame = +1
Query: 1 GVSTDEKVAKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGL 60
G S + A F F ELA AT++F+ +GEGGFGKVYKG + + VA+KQL +G
Sbjct: 382 GSSNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTT-GEAVAVKQLSHDGR 558
Query: 61 QGTREFAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPL 120
QG +EF +EVL LSL H NLV+L+G+C +G+QRLLVYEYMP+GSLE+HL +L K+PL
Sbjct: 559 QGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPL 738
Query: 121 DWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGD 180
+W+TRM++A G A+GLEYLH PPVIYRDLK +NILL ++++PKLSDFGLAK+GP+GD
Sbjct: 739 NWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGD 918
Query: 181 MTHVSTRVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLV 240
THVSTRVMGTYGYCAP+YAM+G+LT KSDIYS GV LLEL+TGR+A D S+ EQNLV
Sbjct: 919 NTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLV 1098
Query: 241 AWAYPLFKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDF 300
+WA P F ++R+F MVDPLL+G++P+R L+QA+A+ AMC++EQ RP+I D+V AL++
Sbjct: 1099SWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEY 1278
Query: 301 IASHKYDPQVH 311
+AS + P+ H
Sbjct: 1279LAS-QGSPEAH 1308
>AV423294
Length = 459
Score = 278 bits (711), Expect = 9e-76
Identities = 128/152 (84%), Positives = 145/152 (95%)
Frame = +3
Query: 32 EGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAVEVLTLSLAEHPNLVKLLGFCAEG 91
EGGFGKVY+G+I++INQ VAIKQL+P+GLQG REF VEVLTLSLA+HPNLVKL+GFCAEG
Sbjct: 3 EGGFGKVYRGFIERINQVVAIKQLNPHGLQGIREFVVEVLTLSLADHPNLVKLIGFCAEG 182
Query: 92 EQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRIAAGVAKGLEYLHDEMKPPVIYRD 151
EQRLLVYEYMPLGSLE+HLHD+ PGKKPLDWNTRM+IAAG A+GLEYLH++MKPPVIYRD
Sbjct: 183 EQRLLVYEYMPLGSLESHLHDILPGKKPLDWNTRMKIAAGAARGLEYLHNKMKPPVIYRD 362
Query: 152 LKCSNILLGDDYHPKLSDFGLAKVGPIGDMTH 183
+KCSN+L+GD YHPKLSDFGLAKVGPIGD TH
Sbjct: 363 IKCSNLLIGDGYHPKLSDFGLAKVGPIGDKTH 458
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 241 bits (615), Expect = 1e-64
Identities = 131/292 (44%), Positives = 187/292 (63%), Gaps = 6/292 (2%)
Frame = +2
Query: 13 TFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTR-EFAVEVL 71
+ DEL T +F +GEG +G+VY + N VA+K+LD + T EF +V
Sbjct: 326 SLDELKEKTDNFGSKALIGEGSYGRVYYATLNDGNA-VAVKKLDVSSEPETNNEFLTQVS 502
Query: 72 TLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLP--PGKKP---LDWNTRM 126
+S ++ N V+L G+C EG R+L YE+ +GSL + LH G +P LDW R+
Sbjct: 503 MVSRLKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLDWIQRV 682
Query: 127 RIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVST 186
RIA A+GLEYLH++++P +I+RD++ SN+L+ +DY K++DF L+ P ST
Sbjct: 683 RIAVDAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDMAARLHST 862
Query: 187 RVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPL 246
RV+GT+GY AP+YAMTGQLT KSD+YS GV LLEL+TGRK D + P +Q+LV WA P
Sbjct: 863 RVLGTFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQSLVTWATPR 1042
Query: 247 FKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAAL 298
E K + VDP L+G+YP +G+ + AVAA+CV+ ++ RP ++ VV AL
Sbjct: 1043LSED-KVKQCVDPKLKGEYPPKGVAKLAAVAALCVQYEAEFRPNMSIVVKAL 1195
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 225 bits (574), Expect = 7e-60
Identities = 122/288 (42%), Positives = 176/288 (60%)
Frame = +3
Query: 12 FTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAVEVL 71
FT + + AT+ ++ +GEGGFG VY+G + Q VA+K QGTREF E+
Sbjct: 2133 FTLEYIEVATERYKT--LIGEGGFGSVYRGTLND-GQEVAVKVRSSTSTQGTREFDNELN 2303
Query: 72 TLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRIAAG 131
LS +H NLV LLG+C E +Q++LVY +M GSL++ L+ P +K LDW TR+ IA G
Sbjct: 2304 LLSAIQHENLVPLLGYCNESDQQILVYPFMSNGSLQDRLYGEPAKRKILDWPTRLSIALG 2483
Query: 132 VAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRVMGT 191
A+GL YLH VI+RD+K SNILL K++DFG +K P ++VS V GT
Sbjct: 2484 AARGLAYLHTFPGRSVIHRDIKSSNILLDHSMCAKVADFGFSKYAPQEGDSYVSLEVRGT 2663
Query: 192 YGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFKEQR 251
GY P+Y T QL+ KSD++S GV LLE+++GR+ + +P E +LV WA P +
Sbjct: 2664 AGYLDPEYYKTQQLSEKSDVFSFGVVLLEIVSGREPLNIKRPRTEWSLVEWATPYIRGS- 2840
Query: 252 KFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALD 299
K ++VDP ++G Y A +++ + VA C+E S+ RP + +V L+
Sbjct: 2841 KVDEIVDPGIKGGYHAEAMWRVVEVALQCLEPFSTYRPSMVAIVRELE 2984
>CN825263
Length = 663
Score = 224 bits (570), Expect = 2e-59
Identities = 115/213 (53%), Positives = 143/213 (66%)
Frame = +1
Query: 34 GFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAVEVLTLSLAEHPNLVKLLGFCAEGEQ 93
GFG VYKG + + VA+K L + +G REF EV LS H NLVKL+G C E +
Sbjct: 1 GFGLVYKGILND-GRDVAVKILKRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQT 177
Query: 94 RLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLK 153
R L+YE +P GS+E+HLH PLDWN RM+IA G A+GL YLH++ P VI+RD K
Sbjct: 178 RCLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRDFK 357
Query: 154 CSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRVMGTYGYCAPDYAMTGQLTSKSDIYS 213
SNILL D+ PK+SDFGLA+ H+ST VMGT+GY AP+YAMTG L KSD+YS
Sbjct: 358 SSNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSDVYS 537
Query: 214 LGVALLELITGRKAFDPSKPAKEQNLVAWAYPL 246
GV LLEL+TG K D S+P ++NLV WA P+
Sbjct: 538 YGVVLLELLTGTKPVDLSQPPGQENLVTWARPI 636
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 220 bits (560), Expect = 3e-58
Identities = 115/256 (44%), Positives = 165/256 (63%)
Frame = +1
Query: 10 KIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAVE 69
++F+ EL +AT +F D +GEGGFG VY G + +Q +A+K+L + EFAVE
Sbjct: 22 RVFSLKELHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQ-IAVKRLKVWSNKADMEFAVE 198
Query: 70 VLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRIA 129
V L+ H NL+ L G+CAEG++RL+VY+YMP SL +HLH + LDWN RM IA
Sbjct: 199 VEILARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECLLDWNRRMNIA 378
Query: 130 AGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRVM 189
G A+G+ YLH + P +I+RD+K SN+LL D+ +++DFG AK+ P G THV+TRV
Sbjct: 379 IGSAEGIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPDG-ATHVTTRVK 555
Query: 190 GTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFKE 249
GT GY AP+YAM G+ D++S G+ LLEL +G+K + ++++ WA PL
Sbjct: 556 GTLGYLAPEYAMLGKANECCDVFSFGILLLELASGKKPLEKLSSTVKRSINDWALPL-AC 732
Query: 250 QRKFSKMVDPLLEGQY 265
+KF++ DP L G+Y
Sbjct: 733 AKKFTEFADPRLNGEY 780
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 209 bits (533), Expect = 4e-55
Identities = 110/237 (46%), Positives = 152/237 (63%)
Frame = +2
Query: 10 KIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAVE 69
+IFT+ EL AAT F D +GEGGFG VY G Q +A+K+L + EFAVE
Sbjct: 260 RIFTYKELHAATGGFSDDNKLGEGGFGSVYWGRTSDGLQ-IAVKKLKAMNSKAEMEFAVE 436
Query: 70 VLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRIA 129
V L H NL+ L G+C +QRL+VY+YMP SL +HLH + L+W RM+IA
Sbjct: 437 VEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQFAVEVQLNWQKRMKIA 616
Query: 130 AGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRVM 189
G A+G+ YLH E+ P +I+RD+K SN+LL D+ P ++DFG AK+ P G ++H++TRV
Sbjct: 617 IGSAEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPLVADFGFAKLIPEG-VSHMTTRVK 793
Query: 190 GTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPL 246
GT GY AP+YAM G+++ D+YS G+ LLEL+TGRK + ++ + WA PL
Sbjct: 794 GTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGVKRTITEWAEPL 964
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 204 bits (520), Expect = 1e-53
Identities = 112/271 (41%), Positives = 159/271 (58%)
Frame = +3
Query: 30 VGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAVEVLTLSLAEHPNLVKLLGFCA 89
+G GGFGKVY G + + VAIK+ +P QG EF E+ LS H +LV L+G+C
Sbjct: 15 LGVGGFGKVYYGEVDGGTK-VAIKRGNPLSEQGVHEFQTEIEMLSKLRHRHLVSLIGYCE 191
Query: 90 EGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRIAAGVAKGLEYLHDEMKPPVIY 149
E + +LVY++M G+L HL+ K PL W R+ I G A+GL YLH K +I+
Sbjct: 192 ENTEMILVYDHMAYGTLREHLYKTQ--KPPLPWKQRLEICIGAARGLHYLHTGAKYTIIH 365
Query: 150 RDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRVMGTYGYCAPDYAMTGQLTSKS 209
RD+K +NILL + + K+SDFGL+K GP D THVST V G++GY P+Y QLT KS
Sbjct: 366 RDVKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVKGSFGYLDPEYFRRQQLTDKS 545
Query: 210 DIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFKEQRKFSKMVDPLLEGQYPARG 269
D+YS GV L E++ R A +PS ++ +L WA + + +++DP L+G+
Sbjct: 546 DVYSFGVVLFEILCARPALNPSLAKEQVSLAEWASHCY-NKGILDQILDPYLKGKIAPEC 722
Query: 270 LYQALAVAAMCVEEQSSMRPVIADVVAALDF 300
+ A CV +Q RP + DV+ L+F
Sbjct: 723 FKKFAETAMKCVSDQGIERPSMGDVLWNLEF 815
>TC19526 homologue to UP|AAQ96340 (AAQ96340) Protein kinase-like protein,
partial (37%)
Length = 432
Score = 197 bits (501), Expect = 2e-51
Identities = 95/133 (71%), Positives = 111/133 (83%)
Frame = +1
Query: 9 AKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAV 68
A+ FTF ELAAATK+FR +C +GEGGFG+VYKG ++ Q VA+KQLD NGLQG REF V
Sbjct: 43 AQTFTFRELAAATKNFRPECLLGEGGFGRVYKGRLESTGQVVAVKQLDRNGLQGNREFLV 222
Query: 69 EVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRI 128
EVL LSL HPNLV L+G+CA+G+QRLLVYE+MPLGSLE+HLH K+PLDWNTRM+I
Sbjct: 223 EVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHG---DKEPLDWNTRMKI 393
Query: 129 AAGVAKGLEYLHD 141
AAG AKGLEYLHD
Sbjct: 394 AAGAAKGLEYLHD 432
>AI967315
Length = 1308
Score = 192 bits (488), Expect = 6e-50
Identities = 109/292 (37%), Positives = 172/292 (58%), Gaps = 2/292 (0%)
Frame = +1
Query: 10 KIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPN--GLQGTREFA 67
K F+++EL AT F + VG+GG+ +VYKG ++ ++ +A+K+L + +EF
Sbjct: 106 KCFSYEELFHATNGFSSENMVGKGGYAEVYKGRLESGDE-IAVKRLTRTCRDERKEKEFL 282
Query: 68 VEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMR 127
E+ T+ H N++ LLG C + LV+E +GS+ + +HD PLDW TR +
Sbjct: 283 TEIGTIGHVCHSNVMPLLGCCIDNGL-YLVFELSTVGSVASLIHD--EKMAPLDWKTRYK 453
Query: 128 IAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTR 187
I G A+GL YLH + +I+RD+K SNILL +D+ P++SDFGLAK P H
Sbjct: 454 IVLGTARGLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPSQWTHHSIAP 633
Query: 188 VMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLF 247
+ GT+G+ AP+Y M G + K+D+++ GV LLE+I+GRK D S Q+L WA P+
Sbjct: 634 IEGTFGHLAPEYYMHGVVDEKTDVFAFGVFLLEVISGRKPVDGS----HQSLHTWAKPIL 801
Query: 248 KEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALD 299
+ + K+VDP LEG Y + A++C+ S+ RP +++V+ ++
Sbjct: 802 SKW-EIEKLVDPRLEGCYDVTQFNRVAFAASLCIRASSTWRPTMSEVLEVME 954
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 191 bits (485), Expect = 1e-49
Identities = 114/301 (37%), Positives = 177/301 (57%), Gaps = 9/301 (2%)
Frame = +1
Query: 12 FTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAVEVL 71
F++ ELA AT +F +D +G+GGFG VY Y + + AIK++D +Q + EF E+
Sbjct: 1051 FSYQELAKATNNFSLDNKIGQGGFGAVY--YAELRGKKTAIKKMD---VQASTEFLCELK 1215
Query: 72 TLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRIAAG 131
L+ H NLV+L+G+C EG LVYE++ G+L +LH GK+PL W++R++IA
Sbjct: 1216 VLTHVHHLNLVRLIGYCVEGSL-FLVYEHIDNGNLGQYLHG--SGKEPLPWSSRVQIALD 1386
Query: 132 VAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRVMGT 191
A+GLEY+H+ P I+RD+K +NIL+ + K++DFGL K+ +G+ T + TR++GT
Sbjct: 1387 AARGLEYIHEHTVPVYIHRDVKSANILIDKNLRGKVADFGLTKLIEVGNST-LQTRLVGT 1563
Query: 192 YGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAF--DPSKPAKEQNLVAWAYPLFKE 249
+GY P+YA G ++ K D+Y+ GV L ELI+ + A A+ + LVA LF+E
Sbjct: 1564 FGYMPPEYAQYGDISPKIDVYAFGVVLFELISAKNAVLKTGELVAESKGLVA----LFEE 1731
Query: 250 QRKFS-------KMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIA 302
S K+VDP L YP + + + C + +RP + +V AL ++
Sbjct: 1732 ALNKSDPCDALRKLVDPRLGENYPIDSVLKIAQLGRACTRDNPLLRPSMRSLVVALMTLS 1911
Query: 303 S 303
S
Sbjct: 1912 S 1914
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 190 bits (483), Expect = 2e-49
Identities = 115/296 (38%), Positives = 173/296 (57%), Gaps = 1/296 (0%)
Frame = +1
Query: 5 DEKVAKIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTR 64
D +A IF F +++AT F + +GEGGFG VYKG + Q +A+K+L QG
Sbjct: 1615 DIDLATIFDFSTISSATNHFSLSNKLGEGGFGPVYKGLLAN-GQEIAVKRLSNTSGQGME 1791
Query: 65 EFAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNT 124
EF E+ ++ +H NLVKL G ++ + M + L D K +DWN
Sbjct: 1792 EFKNEIKLIARLQHRNLVKLFGCSVHQDENSHANKKMKI------LLDSTRSKL-VDWNK 1950
Query: 125 RMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHV 184
R++I G+A+GL YLH + + +I+RDLK SNILL D+ +PK+SDFGLA++ IGD
Sbjct: 1951 RLQIIDGIARGLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIF-IGDQVEA 2127
Query: 185 ST-RVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWA 243
T RVMGTYGY P+YA+ G + KSD++S GV +LE+I+G+K P NL++ A
Sbjct: 2128 RTKRVMGTYGYMPPEYAVHGSFSIKSDVFSFGVIVLEIISGKKIGRFYDPHHHLNLLSHA 2307
Query: 244 YPLFKEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALD 299
+ L+ E+R ++VD LL+ + + + VA +CV+ + RP + +V L+
Sbjct: 2308 WRLWIEERPL-ELVDELLDDPVIPTEILRYIHVALLCVQRRPENRPDMLSIVLMLN 2472
>AV765034
Length = 529
Score = 189 bits (481), Expect = 4e-49
Identities = 90/121 (74%), Positives = 107/121 (88%)
Frame = -3
Query: 196 APDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFKEQRKFSK 255
APDYAMTGQLT KSD+YS GV LLE+ITGRKA D +KPAKEQNLVAWA PLF++++KFS
Sbjct: 527 APDYAMTGQLTFKSDVYSFGVVLLEIITGRKAIDHTKPAKEQNLVAWARPLFRDRKKFSL 348
Query: 256 MVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAALDFIASHKYDPQVHPIQS 315
MVDPLLEGQYP RGLYQALA+AAMCV+EQ +MRPVI DVV AL+++A++KYDPQ ++S
Sbjct: 347 MVDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIVDVVTALNYLATNKYDPQNQTVES 168
Query: 316 S 316
S
Sbjct: 167 S 165
>TC9461 similar to UP|O49840 (O49840) Protein kinase (AT2G02800/T20F6.6) ,
partial (37%)
Length = 484
Score = 183 bits (465), Expect = 3e-47
Identities = 91/163 (55%), Positives = 118/163 (71%)
Frame = +3
Query: 100 YMPLGSLENHLHDLPPGKKPLDWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILL 159
+MP GSLENHL G +PL W+ RM++A G A+GL +LH+ K VIYRD K SNILL
Sbjct: 3 FMPKGSLENHL--FRRGPQPLSWSVRMKVAIGAARGLSFLHNA-KSQVIYRDFKASNILL 173
Query: 160 GDDYHPKLSDFGLAKVGPIGDMTHVSTRVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALL 219
+++ KLSDFGLAK GP GD THVST+VMGT GY AP+Y TG+LT+KSD+YS GV LL
Sbjct: 174 DAEFNAKLSDFGLAKAGPTGDRTHVSTQVMGTQGYAAPEYVATGRLTAKSDVYSFGVVLL 353
Query: 220 ELITGRKAFDPSKPAKEQNLVAWAYPLFKEQRKFSKMVDPLLE 262
EL++GR+A D + +QNLV WA P ++R+ +++D LE
Sbjct: 354 ELLSGRRAVDKTIAGVDQNLVDWARPYLGDKRRLFRIMDSKLE 482
>TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P14.15
{Arabidopsis thaliana;}, partial (37%)
Length = 1079
Score = 178 bits (451), Expect = 1e-45
Identities = 99/227 (43%), Positives = 141/227 (61%), Gaps = 1/227 (0%)
Frame = +3
Query: 69 EVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRI 128
EV L+ +H NLVKLLGF +G +R+L+ EY+P G+L HL L K LD+N R+ I
Sbjct: 6 EVELLAKIDHRNLVKLLGFIDKGNERILITEYVPNGTLREHLDGLRG--KILDFNQRLEI 179
Query: 129 AAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPI-GDMTHVSTR 187
A VA GL YLH + +I+RD+K SNILL + K++DFG A++GP+ GD TH+ST+
Sbjct: 180 AIDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFARLGPVNGDQTHISTK 359
Query: 188 VMGTYGYCAPDYAMTGQLTSKSDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLF 247
V GT GY P+Y T QLT KSD+YS G+ LLE++TGR+ + K A E+ + WA+ +
Sbjct: 360 VKGTVGYLDPEYMKTHQLTPKSDVYSFGILLLEILTGRRPVELKKAADERVTLRWAFRKY 539
Query: 248 KEQRKFSKMVDPLLEGQYPARGLYQALAVAAMCVEEQSSMRPVIADV 294
E +++DPL+E A L + L ++ C + RP + V
Sbjct: 540 NE-GSVVELMDPLMEEAVNADVLMKMLDLSFQCAAPIRTDRPNMKSV 677
>TC12473 similar to UP|Q9M068 (Q9M068) Protein kinase-like protein, partial
(42%)
Length = 946
Score = 176 bits (447), Expect = 4e-45
Identities = 93/180 (51%), Positives = 122/180 (67%), Gaps = 9/180 (5%)
Frame = +3
Query: 10 KIFTFDELAAATKSFRVDCFVGEGGFGKVYKGYI--KKINQ-------FVAIKQLDPNGL 60
K+F+F +L +ATKSF+ D +GEGGFGKVYKG++ KK++ VA+K+L+P +
Sbjct: 414 KVFSFGDLKSATKSFKADALIGEGGFGKVYKGWLDEKKLSPTKPGSGIMVAVKKLNPESM 593
Query: 61 QGTREFAVEVLTLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPL 120
QG E+ E+ L HPNLVKLLG+C + E+ LLVYE+MP GSLENHL + L
Sbjct: 594 QGFHEWQSEINFLGRISHPNLVKLLGYCRDDEEFLLVYEFMPRGSLENHL--FRRNTESL 767
Query: 121 DWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGD 180
WNTR++IA G A+GL +LH K VIYRD K SNILL +Y+ K+S FGL K GP G+
Sbjct: 768 SWNTRLKIAIGAARGLAFLHSLXK-IVIYRDFKASNILLDGNYNAKISXFGLTKFGPSGE 944
>AV417846
Length = 429
Score = 169 bits (428), Expect = 6e-43
Identities = 84/144 (58%), Positives = 103/144 (71%)
Frame = +1
Query: 100 YMPLGSLENHLHDLPPGKKPLDWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILL 159
+MP GS+ENHL +P W+ R++IA G AKGL +LH +P VIYRD K SNILL
Sbjct: 1 FMPKGSMENHLFRRGSYFQPFSWSLRLKIALGAAKGLAFLHST-EPKVIYRDFKTSNILL 177
Query: 160 GDDYHPKLSDFGLAKVGPIGDMTHVSTRVMGTYGYCAPDYAMTGQLTSKSDIYSLGVALL 219
Y KLSDFGLA+ GP+GD +HVSTRVMGT GY AP+Y TG LT+ SD+YS GV LL
Sbjct: 178 DTKYSAKLSDFGLARDGPVGDKSHVSTRVMGTRGYAAPEYLATGHLTANSDVYSFGVVLL 357
Query: 220 ELITGRKAFDPSKPAKEQNLVAWA 243
E+I+GR+A D ++ A E NLV WA
Sbjct: 358 EIISGRRAIDKNQLAGEHNLVEWA 429
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 167 bits (423), Expect = 2e-42
Identities = 106/276 (38%), Positives = 149/276 (53%), Gaps = 7/276 (2%)
Frame = +2
Query: 30 VGEGGFGKVYKGYIKKINQFVAIKQLDPNGL-QGTREFAVEVLTLSLAEHPNLVKLLGFC 88
+G+GG G VY+G + VAIK+L G + F E+ TL H N+++LLG+
Sbjct: 2210 IGKGGAGIVYRGSMPNGTD-VAIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRLLGYV 2386
Query: 89 AEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRIAAGVAKGLEYLHDEMKPPVI 148
+ + LL+YEYMP GSL LH G L W R +IA A+GL Y+H + P +I
Sbjct: 2387 SNKDTNLLLYEYMPNGSLGEWLHGAKGGH--LRWEMRYKIAVEAARGLCYMHHDCSPLII 2560
Query: 149 YRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRVMGTYGYCAPDYAMTGQLTSK 208
+RD+K +NILL D+ ++DFGLAK + + + G+YGY AP+YA T ++ K
Sbjct: 2561 HRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEK 2740
Query: 209 SDIYSLGVALLELITGRKAFDPSKPAKEQNLVAWAYPLFKEQRKFS------KMVDPLLE 262
SD+YS GV LLELI GRK + ++V W E + S +VDP L
Sbjct: 2741 SDVYSFGVVLLELIIGRKPV--GEFGDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRLS 2914
Query: 263 GQYPARGLYQALAVAAMCVEEQSSMRPVIADVVAAL 298
G YP + +A MCV+E RP + +VV L
Sbjct: 2915 G-YPLTSVIHMFNIAMMCVKEMGPARPTMREVVHML 3019
>BP032158
Length = 555
Score = 166 bits (420), Expect = 5e-42
Identities = 87/182 (47%), Positives = 115/182 (62%)
Frame = +2
Query: 12 FTFDELAAATKSFRVDCFVGEGGFGKVYKGYIKKINQFVAIKQLDPNGLQGTREFAVEVL 71
F+ ++ AAT +F +GEGGFG VYKG + + +A+KQL QG REF E+
Sbjct: 14 FSLRQIKAATNNFDPANKIGEGGFGPVYKGVLSE-GDVIAVKQLSSKSKQGNREFINEIG 190
Query: 72 TLSLAEHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLPPGKKPLDWNTRMRIAAG 131
+S +HPNLVKL G C EG Q LLVYEYM SL L K L+W TRM+I G
Sbjct: 191 MISALQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGNEEQKLNLNWRTRMKICVG 370
Query: 132 VAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFGLAKVGPIGDMTHVSTRVMGT 191
+AKGL YLH+E + +++RD+K +N+LL D K+SDFGLAK+ + TH+STR+ GT
Sbjct: 371 IAKGLAYLHEESRLKIVHRDIKATNVLLDKDLKAKISDFGLAKLDE-EENTHISTRIAGT 547
Query: 192 YG 193
G
Sbjct: 548 IG 553
>AV767377
Length = 444
Score = 165 bits (418), Expect = 8e-42
Identities = 77/104 (74%), Positives = 86/104 (82%)
Frame = +1
Query: 112 DLPPGKKPLDWNTRMRIAAGVAKGLEYLHDEMKPPVIYRDLKCSNILLGDDYHPKLSDFG 171
+L P +KPLDW TRM+IA G AKGLEYLHD PPVIYRD K SNILL DD++PKL DFG
Sbjct: 133 ELTPERKPLDWKTRMKIAEGAAKGLEYLHDVANPPVIYRDFKASNILLDDDFNPKLFDFG 312
Query: 172 LAKVGPIGDMTHVSTRVMGTYGYCAPDYAMTGQLTSKSDIYSLG 215
LAK+GP GD THVSTR MGTYGY AP+YA TGQLT+KSD+YS G
Sbjct: 313 LAKLGPTGDKTHVSTRGMGTYGYWAPEYASTGQLTTKSDVYSFG 444
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,963,037
Number of Sequences: 28460
Number of extensions: 63941
Number of successful extensions: 742
Number of sequences better than 10.0: 303
Number of HSP's better than 10.0 without gapping: 551
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 557
length of query: 317
length of database: 4,897,600
effective HSP length: 90
effective length of query: 227
effective length of database: 2,336,200
effective search space: 530317400
effective search space used: 530317400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Medicago: description of AC149490.2


