

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149489.2 - phase: 0
(365 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
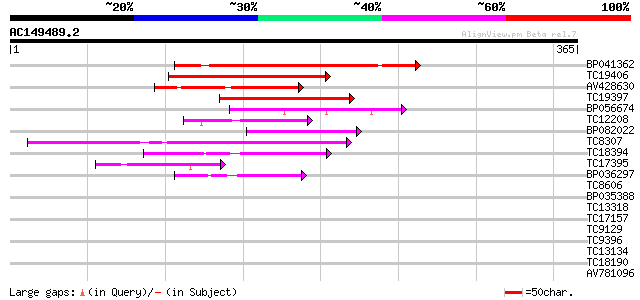
Score E
Sequences producing significant alignments: (bits) Value
BP041362 161 1e-40
TC19406 similar to UP|Q9FJS2 (Q9FJS2) Homeobox protein, partial ... 115 2e-26
AV428630 91 3e-19
TC19397 similar to PIR|S47137|S47137 homeotic protein Athb-7 - A... 69 1e-12
BP056674 60 6e-10
TC12208 similar to UP|Q8LC03 (Q8LC03) Homeobox gene 13 protein, ... 55 1e-08
BP082022 55 1e-08
TC8307 weakly similar to UP|Q9SP47 (Q9SP47) Homeodomain-leucine ... 55 2e-08
TC18394 similar to UP|O23208 (O23208) Homeodomain protein, parti... 50 5e-07
TC17395 similar to UP|Q9LN83 (Q9LN83) T12C24.17, partial (37%) 45 3e-05
BP036297 44 3e-05
TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%) 39 0.001
BP035388 38 0.003
TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (... 38 0.003
TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly pr... 37 0.004
TC9129 homologue to UP|Q8LLE2 (Q8LLE2) BEL1-related homeotic pro... 37 0.005
TC9396 similar to UP|Q9ZRB9 (Q9ZRB9) Homeobox 1 protein (Fragmen... 37 0.005
TC13134 similar to UP|O80409 (O80409) Pharbitis kntted-like gene... 37 0.007
TC18190 weakly similar to UP|O24569 (O24569) HOX1B protein, part... 36 0.009
AV781096 36 0.012
>BP041362
Length = 560
Score = 161 bits (408), Expect = 1e-40
Identities = 81/158 (51%), Positives = 105/158 (66%)
Frame = +3
Query: 107 EYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDE 166
E E D E+ EG G ++ + K+K+YHRHT QI+ MEA FKE PHPD+
Sbjct: 96 EDEFDSATKSGSENHEG-----ASGEDQEPRAKKKRYHRHTQHQIQEMEAFFKECPHPDD 260
Query: 167 KQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINK 226
KQR++LS++LGL P QVKFWFQN+RTQ+K ERHEN+ L++E EKLR N RE ++
Sbjct: 261 KQRKELSRELGLEPLQVKFWFQNKRTQMKTQHERHENTSLRTENEKLRADNMRYREALSN 440
Query: 227 ACCPNCGVPTTNRDGTMATEEQQLRIENAKLKAEVERL 264
A CPNCG PT G M+ +E LR ENA+L+ E ER+
Sbjct: 441 ASCPNCGGPTA--XGEMSFDEHHLRXENARLREEXERI 548
>TC19406 similar to UP|Q9FJS2 (Q9FJS2) Homeobox protein, partial (10%)
Length = 546
Score = 115 bits (287), Expect = 2e-26
Identities = 56/104 (53%), Positives = 72/104 (68%)
Frame = +1
Query: 103 KSIDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESP 162
K D G + E + G + E D G+ ++ K+K+YHRHT+ QI+ MEALFKE P
Sbjct: 235 KEEDSMLGGKEEMESGSESEQVEDKSGNEQEITEQPKKKRYHRHTARQIQEMEALFKECP 414
Query: 163 HPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLL 206
HPD+KQR +LS LGL PRQVKFWFQNRRTQ+KA Q+R +N +L
Sbjct: 415 HPDDKQRMKLSHDLGLKPRQVKFWFQNRRTQMKAQQDRSDNVIL 546
>AV428630
Length = 342
Score = 90.9 bits (224), Expect = 3e-19
Identities = 45/96 (46%), Positives = 59/96 (60%)
Frame = +1
Query: 94 SEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRV 153
S+ + + + DE+ D E+ EG G NKKK+ YHRHT QI+
Sbjct: 70 SDIAAAVRIRGEDEF--DSATKSGSENQEGGASGGDQDPRPNKKKR---YHRHTQHQIQE 234
Query: 154 MEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQN 189
ME+ FKE PHPD+KQR++LS++LGL P QVKFWFQN
Sbjct: 235 MESFFKECPHPDDKQRKELSRELGLEPLQVKFWFQN 342
>TC19397 similar to PIR|S47137|S47137 homeotic protein Athb-7 - Arabidopsis
thaliana {Arabidopsis thaliana;}, partial (35%)
Length = 723
Score = 69.3 bits (168), Expect = 1e-12
Identities = 32/87 (36%), Positives = 54/87 (61%)
Frame = +2
Query: 136 KKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIK 195
+KK+ K R + EQI+ +E++F+ + +++ QL+++LGL PRQV WFQN+R + K
Sbjct: 239 RKKRNKNARRFSDEQIKSLESMFESESRLEPRKKLQLARELGLQPRQVAIWFQNKRARWK 418
Query: 196 AIQERHENSLLKSEIEKLREKNKTLRE 222
+ Q E S+L S L K + L++
Sbjct: 419 SKQLEREYSMLSSNYNNLASKYEVLKK 499
>BP056674
Length = 566
Score = 60.1 bits (144), Expect = 6e-10
Identities = 38/134 (28%), Positives = 62/134 (45%), Gaps = 20/134 (14%)
Frame = +1
Query: 142 KYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQ----LGLAPRQVKFWFQNRRTQIKAI 197
KY R+T+ Q+ +E ++ E P P +RQQL ++ + P+Q+K WFQNRR + K
Sbjct: 118 KYVRYTAGQVEALERVYTECPKPSSLRRQQLIRECPVLANVEPKQIKVWFQNRRCREKQR 297
Query: 198 QERHE----NSLLKSEIEKLREKNKTLRETINKACCPN------------CGVPTTNRDG 241
+E N L + + L E+N L++ +++ C N G N D
Sbjct: 298 KEASRLQAVNRKLNAMNKLLMEENDRLQKQVSQLVCENGFMRQQLQAPSAAGTTDGNGDS 477
Query: 242 TMATEEQQLRIENA 255
T +R N+
Sbjct: 478 VATTSRNSMRDANS 519
>TC12208 similar to UP|Q8LC03 (Q8LC03) Homeobox gene 13 protein, partial
(24%)
Length = 660
Score = 55.5 bits (132), Expect = 1e-08
Identities = 33/87 (37%), Positives = 47/87 (53%), Gaps = 4/87 (4%)
Frame = +1
Query: 113 LEDDEGEDDE----GDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQ 168
LE + + DE GD D DG +KKKR EQ++ +E F+ + ++
Sbjct: 400 LEMNNNKCDEVLHGGDDDLSDDGSQLGEKKKRLSL-----EQVKALERSFELGNKLEPER 564
Query: 169 RQQLSKQLGLAPRQVKFWFQNRRTQIK 195
+ QL+K LGL PRQ+ WFQNRR + K
Sbjct: 565 KMQLAKALGLQPRQIAIWFQNRRARWK 645
>BP082022
Length = 455
Score = 55.5 bits (132), Expect = 1e-08
Identities = 28/74 (37%), Positives = 44/74 (58%)
Frame = -3
Query: 153 VMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEK 212
++E FK+ + KQ+Q L++QL L PRQV+ WFQNRR + K Q + LK E
Sbjct: 447 MLEESFKQHSTLNPKQKQALARQLNLRPRQVEVWFQNRRARTKLKQTEVDCDFLKKCCET 268
Query: 213 LREKNKTLRETINK 226
L ++N L++ + +
Sbjct: 267 LTDENMRLQKELQE 226
>TC8307 weakly similar to UP|Q9SP47 (Q9SP47) Homeodomain-leucine zipper
protein 57, partial (51%)
Length = 1629
Score = 55.1 bits (131), Expect = 2e-08
Identities = 53/210 (25%), Positives = 87/210 (41%), Gaps = 1/210 (0%)
Frame = +1
Query: 12 HNTLLSSPTLSMCADMSNNNP-PSTSKAKDFFPSPALSLSLAGIFRHGGGAAAEGEGGSI 70
H+ LL L + + + P P A + LS +AG GG+AA
Sbjct: 271 HHQLLPPQVLDLLPLLRSTKPSPMAR*ALPWIKLLFLSDFMAGGRVFSGGSAAPANVSDT 450
Query: 71 SNMEVEEGEEGSTIGGERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGD 130
S + + + S S+ + G S+S+ + ++L D D +GD
Sbjct: 451 SLLLQNQPPDSSLF-----LSTSASFLG---SRSMVSFADNKLGQTRSFFSAFDLDENGD 606
Query: 131 GVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNR 190
V + +K R + +Q++ +E F + ++ +++K LGL PRQV WFQNR
Sbjct: 607 EVMDEYFHQSEKKRRLSVDQVQFLEKSF*VDNKLEPDRKTKIAKDLGLQPRQVAIWFQNR 786
Query: 191 RTQIKAIQERHENSLLKSEIEKLREKNKTL 220
R + K Q + L S E L+ L
Sbjct: 787 RARWKTKQLEKDYDSLHSSFESLKSNYDNL 876
>TC18394 similar to UP|O23208 (O23208) Homeodomain protein, partial (46%)
Length = 846
Score = 50.4 bits (119), Expect = 5e-07
Identities = 33/121 (27%), Positives = 60/121 (49%)
Frame = +2
Query: 87 ERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRH 146
E++ +S Y ++ I + +G++ + + + G G+ NKK+K +
Sbjct: 29 EQMMLMSQLYPAATYNQIISQQQGEKKQPRRRRNKKNRG-GENGSAEANKKRKLSE---- 193
Query: 147 TSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLL 206
EQ+ ++E F + +++ +L+ +L L PRQV WFQNRR + K + E S L
Sbjct: 194 --EQVNLLEQNFGHEHKLESERKDKLAAELCLDPRQVAVWFQNRRARWKNKKLEEEYSNL 367
Query: 207 K 207
K
Sbjct: 368 K 370
>TC17395 similar to UP|Q9LN83 (Q9LN83) T12C24.17, partial (37%)
Length = 570
Score = 44.7 bits (104), Expect = 3e-05
Identities = 30/91 (32%), Positives = 40/91 (42%), Gaps = 7/91 (7%)
Frame = -3
Query: 56 RHGGGAAAEGEGGSISNMEVEEGEEGSTIGGERVEEISSEYSGPAKSKSIDEYEGDELED 115
R + A E GS++ E G+E IGGE E+ E + + E GDE +
Sbjct: 313 RDDAASGAVKEHGSVTE---EHGDEEVEIGGEAEGEVGEENTDGGGEDVLGEATGDEAPE 143
Query: 116 -------DEGEDDEGDGDGDGDGVNKNKKKK 139
DE E++ DGDGD DG K K
Sbjct: 142 EAERNGGDEAEEEAEDGDGDADGCGHGIKVK 50
>BP036297
Length = 569
Score = 44.3 bits (103), Expect = 3e-05
Identities = 27/85 (31%), Positives = 48/85 (55%)
Frame = +3
Query: 107 EYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDE 166
E E D L + G ++E + +G NK K+K + ++ Q+ ++E + +P E
Sbjct: 93 ECELDFLAMECGSEEENKKNTP-EGENKVKRKMK------SASQLELLEKTYAVEAYPGE 251
Query: 167 KQRQQLSKQLGLAPRQVKFWFQNRR 191
R +LS++LGL+ RQ++ WF +RR
Sbjct: 252 VLRAELSEKLGLSDRQLQMWFCHRR 326
>TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%)
Length = 1008
Score = 39.3 bits (90), Expect = 0.001
Identities = 34/101 (33%), Positives = 49/101 (47%), Gaps = 16/101 (15%)
Frame = -2
Query: 58 GGGAAAEGEGGSISNMEVEEGEEGSTIGGERVEEISSEYSGPAKSKSIDEY---EGDELE 114
GGGA AE +++ V+ + + G + +++ G KS+S D+ EG++
Sbjct: 779 GGGALAEVVVTLVASAVVKATLLAADLCGLLLINMATT-KGETKSQSEDKQHDAEGEDGN 603
Query: 115 DDEG-EDDEGDG------------DGDGDGVNKNKKKKRKK 142
DDEG EDD+GDG DG G G N N K KK
Sbjct: 602 DDEGDEDDDGDGPFGEGEEDLSSEDGAGYGNNSNNKSNSKK 480
Score = 37.4 bits (85), Expect = 0.004
Identities = 25/94 (26%), Positives = 39/94 (40%), Gaps = 10/94 (10%)
Frame = -2
Query: 59 GGAAAEGEGGSISNMEVEEGEEGSTIGGERVEEISSEYSGPAKSKSIDEYEGDELEDDEG 118
G ++ + EG G+ G E E + G + D+ E D+ EDD G
Sbjct: 527 GAGYGNNSNNKSNSKKAPEGGPGAGAGPEENGEDEEDEDGEDQEDDDDDDEDDDEEDDGG 348
Query: 119 EDDEGDGDGDGDGVNKNK----------KKKRKK 142
ED+E +G + D ++ + KKRKK
Sbjct: 347 EDEEEEGVEEEDNEDEEEDEEDEEALQPPKKRKK 246
Score = 35.0 bits (79), Expect = 0.020
Identities = 30/120 (25%), Positives = 53/120 (44%), Gaps = 25/120 (20%)
Frame = -2
Query: 63 AEGEGGSISNMEVEEGEEGSTIGGERVEEISSEY-------------------------S 97
AEGE G+ + E +E ++G GE E++SSE +
Sbjct: 623 AEGEDGN--DDEGDEDDDGDGPFGEGEEDLSSEDGAGYGNNSNNKSNSKKAPEGGPGAGA 450
Query: 98 GPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEAL 157
GP ++ +E E E ++D+ +DDE D + D G ++ ++ ++ + E EAL
Sbjct: 449 GPEENGEDEEDEDGEDQEDDDDDDEDDDEEDDGGEDEEEEGVEEEDNEDEEEDEEDEEAL 270
>BP035388
Length = 557
Score = 37.7 bits (86), Expect = 0.003
Identities = 27/104 (25%), Positives = 53/104 (50%), Gaps = 9/104 (8%)
Frame = +3
Query: 113 LEDDEGEDDEGDGDGDGD----GVNKNKKKKRKKYHRHTSEQIRVMEALFKE---SPHPD 165
+ED +G+ D + + G + K + + EQI ++E++F +P D
Sbjct: 3 MEDHQGQKDPNTKNNNAPSPHHGCSSEKSEPVRSRWTPKPEQILILESIFNSGMVNPPKD 182
Query: 166 EKQR-QQLSKQLG-LAPRQVKFWFQNRRTQIKAIQERHENSLLK 207
E R ++L ++ G + V +WFQNRR++ + Q + + SL++
Sbjct: 183 ETVRIRKLLEKFGTVGDANVFYWFQNRRSRSRRRQRQMQASLVE 314
>TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (SPBC3D6.14C
protein) (Fragment), partial (15%)
Length = 543
Score = 37.7 bits (86), Expect = 0.003
Identities = 16/33 (48%), Positives = 22/33 (66%)
Frame = +3
Query: 99 PAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDG 131
P ++ + +E + DE EDDE EDD+ D D D DG
Sbjct: 234 PVQTDNEEEEDDDEEEDDEDEDDDDDDDDDDDG 332
Score = 29.6 bits (65), Expect = 0.84
Identities = 12/33 (36%), Positives = 19/33 (57%)
Frame = +3
Query: 98 GPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGD 130
GP K D E ++ +++E ++DE D D D D
Sbjct: 222 GPPKPVQTDNEEEEDDDEEEDDEDEDDDDDDDD 320
Score = 26.9 bits (58), Expect = 5.4
Identities = 11/33 (33%), Positives = 21/33 (63%)
Frame = +3
Query: 106 DEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKK 138
D+ +G+E EDD+ +D+E +G + + K+ K
Sbjct: 318 DDDDGEEEEDDDDDDEEEEGSEEVEVEGKDGAK 416
Score = 26.6 bits (57), Expect = 7.1
Identities = 9/23 (39%), Positives = 17/23 (73%)
Frame = +3
Query: 106 DEYEGDELEDDEGEDDEGDGDGD 128
++ + D+ +DD+GE++E D D D
Sbjct: 294 EDDDDDDDDDDDGEEEEDDDDDD 362
>TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly protein
1-like protein 3, partial (81%)
Length = 1298
Score = 37.4 bits (85), Expect = 0.004
Identities = 22/84 (26%), Positives = 39/84 (46%), Gaps = 11/84 (13%)
Frame = +2
Query: 70 ISNMEVEEGEEGSTIGGERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGD--- 126
+ N+ + + GSTI + + S ++G A+ ++ E D+ + DE ED++ D D
Sbjct: 656 LQNLMEHDYDIGSTIRDKIIPHAVSWFTGEAEQSDFEDIEEDDEDGDEDEDEDDDDDEEE 835
Query: 127 --------GDGDGVNKNKKKKRKK 142
D +G K+K K K
Sbjct: 836 EDDDDDDEDDEEGEGKSKSKSGSK 907
>TC9129 homologue to UP|Q8LLE2 (Q8LLE2) BEL1-related homeotic protein 13
(Fragment), partial (22%)
Length = 604
Score = 37.0 bits (84), Expect = 0.005
Identities = 17/60 (28%), Positives = 34/60 (56%), Gaps = 2/60 (3%)
Frame = +2
Query: 162 PHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQI--KAIQERHENSLLKSEIEKLREKNKT 219
P+P + + L++Q GL+ QV WF N R ++ ++E ++ + E + RE+N++
Sbjct: 221 PYPSDADKHLLARQTGLSRNQVSNWFINARVRLWKPMVEEMYQQERKEVECAEERERNQS 400
>TC9396 similar to UP|Q9ZRB9 (Q9ZRB9) Homeobox 1 protein (Fragment),
partial (68%)
Length = 1138
Score = 37.0 bits (84), Expect = 0.005
Identities = 45/193 (23%), Positives = 80/193 (41%), Gaps = 11/193 (5%)
Frame = +1
Query: 59 GGAAAEGEGGSISNMEVEEGEEGSTIGGERVEEISSEYSGPA----KSKSIDEYEGDELE 114
G + EG G ++S+ E ++ E + + ++ S GP +S+ E EL+
Sbjct: 427 GVSPGEGTGATMSDDEDDQAESNANLYESSMDGADSLGFGPLVPTESERSLMERVRHELK 606
Query: 115 DDEGEDDEGDGDGDGDGVNKNKKKKRK-KYHRHTSEQIRVMEALFKESPHPDEKQRQQLS 173
E +G + D + +K+R K T+ ++ P+P E+ + +L
Sbjct: 607 H---ELKQGYKEKIVDIREEILRKRRAGKLPGDTTSLLKAWWQAHSNWPYPTEEDKARLV 777
Query: 174 KQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINK------A 227
++ GL +Q+ WF N+R + H N S + K K+ ET N+ +
Sbjct: 778 QETGLQLKQINNWFINQRK-----RNWHTN---PSSATSTKTKRKSAGETSNQSFM*MNS 933
Query: 228 CCPNCGVPTTNRD 240
C N V N D
Sbjct: 934 C*SNMNVKWWNLD 972
>TC13134 similar to UP|O80409 (O80409) Pharbitis kntted-like gene 2, partial
(29%)
Length = 502
Score = 36.6 bits (83), Expect = 0.007
Identities = 19/73 (26%), Positives = 38/73 (52%)
Frame = +2
Query: 135 NKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQI 194
+KKKK+ K + + + + + P+P E + +L+K GL +Q+ WF N+R +
Sbjct: 35 SKKKKKGKLPKEARQTLLQWWNVHYKWPYPTESDKIELAKATGLDQKQINNWFINQRKRH 214
Query: 195 KAIQERHENSLLK 207
E + S+++
Sbjct: 215 WKPSENMQFSMME 253
>TC18190 weakly similar to UP|O24569 (O24569) HOX1B protein, partial (10%)
Length = 981
Score = 36.2 bits (82), Expect = 0.009
Identities = 17/59 (28%), Positives = 32/59 (53%)
Frame = +3
Query: 133 NKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRR 191
+++KK + R ++ + FK++ +PD ++ L+++LGL QV WF N R
Sbjct: 417 SRDKKPRSSTRQRLGDVVVQRLHKSFKDNQYPDRATKESLAEELGLTFFQVDKWFGNAR 593
>AV781096
Length = 497
Score = 35.8 bits (81), Expect = 0.012
Identities = 14/34 (41%), Positives = 23/34 (67%)
Frame = +3
Query: 142 KYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQ 175
KY R+T+EQ+ +E ++ E P P +RQQL ++
Sbjct: 363 KYVRYTAEQVEALERVYAECPKPSSLRRQQLIRE 464
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.309 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,911,413
Number of Sequences: 28460
Number of extensions: 80101
Number of successful extensions: 1222
Number of sequences better than 10.0: 179
Number of HSP's better than 10.0 without gapping: 890
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1086
length of query: 365
length of database: 4,897,600
effective HSP length: 91
effective length of query: 274
effective length of database: 2,307,740
effective search space: 632320760
effective search space used: 632320760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC149489.2


