

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149489.11 - phase: 0
(526 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
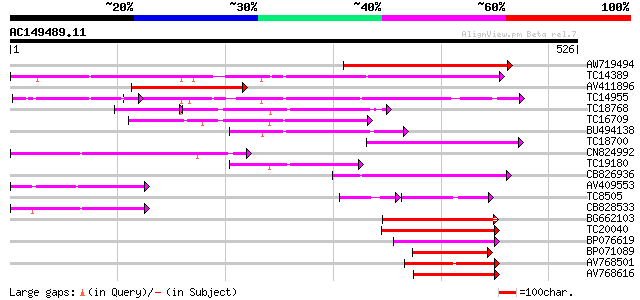
Score E
Sequences producing significant alignments: (bits) Value
AW719494 244 3e-65
TC14389 weakly similar to UP|Q9FNL8 (Q9FNL8) Peptide transporter... 231 2e-61
AV411896 166 9e-42
TC14955 UP|O22305 (O22305) Peptide transporter, complete 138 3e-33
TC18768 107 9e-29
TC16709 weakly similar to PIR|T47573|T47573 peptide transport-li... 120 4e-28
BU494138 114 3e-26
TC18700 similar to UP|Q9LYR6 (Q9LYR6) Peptide transporter-like p... 100 1e-21
CN824992 98 4e-21
TC19180 similar to PIR|T10255|T10255 nitrite transport protein, ... 91 4e-19
CB826936 89 2e-18
AV409553 87 5e-18
TC8505 similar to UP|Q8LG02 (Q8LG02) Nitrate transporter, partia... 65 2e-17
CB828533 83 1e-16
BG662103 82 2e-16
TC20040 similar to UP|Q9FM20 (Q9FM20) Nitrate transporter NTL1, ... 79 2e-15
BP076619 76 2e-14
BP071089 72 2e-13
AV768501 68 4e-12
AV768616 67 5e-12
>AW719494
Length = 473
Score = 244 bits (623), Expect = 3e-65
Identities = 110/157 (70%), Positives = 131/157 (83%)
Frame = +3
Query: 310 LFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQ 369
L F+FLHL+T+A ET+RRK AI+ GY+N+ HGVL MSAMWLAPQLCL GIAE FN IGQ
Sbjct: 3 LLFSFLHLVTSATLETIRRKRAIEAGYINNAHGVLNMSAMWLAPQLCLGGIAEAFNAIGQ 182
Query: 370 NEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHF 429
NEFYY EFP++MS VA+SL GL M GN+VSS V S +E+ T SGG EGWVSDNINKG F
Sbjct: 183 NEFYYTEFPRTMSXVASSLLGLGMAAGNIVSSFVFSAVENITSSGGKEGWVSDNINKGRF 362
Query: 430 DKYYWVIVGINALNLLYYLVCSWAYGPTVDEVSNVSK 466
DKYYWV+ GI+ LNL+YYL+CSWAYGPTVD++S V++
Sbjct: 363 DKYYWVVXGISGLNLVYYLICSWAYGPTVDQISKVAE 473
>TC14389 weakly similar to UP|Q9FNL8 (Q9FNL8) Peptide transporter, partial
(72%)
Length = 1990
Score = 231 bits (589), Expect = 2e-61
Identities = 156/470 (33%), Positives = 240/470 (50%), Gaps = 11/470 (2%)
Frame = +3
Query: 1 MSLLWLTAMIPSARPPACNHPSEG--CESATPGQLAMLFSALILIAIGNGGISCSLA-FG 57
M LL LT +P+ RPP C E C A+P + + AL +IA+G GG +++ G
Sbjct: 246 MCLLTLTVSLPALRPPPCAQGVENQDCPQASPWTRGIFYLALYIIALGAGGTKPNISTMG 425
Query: 58 ADQVNRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLI 117
ADQ + + N L FF+W+ + V+ + T +VYIQD+L W +G+G+P + +
Sbjct: 426 ADQFDEYEPKENTYKLS-FFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLAF 602
Query: 118 STVLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLP--PKTSPEFYHQQ---KDS 172
S ++F + +P Y S T QV VAA K +P PK E ++
Sbjct: 603 SILVFLVGTPFYRHKLPSGSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSR 782
Query: 173 ELVVPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAI 232
+ T LRFL+KA V + W LCTV QVEE K + K++P+ T +
Sbjct: 783 NRIDHTSTLRFLDKAAV----------KTGKTSPWRLCTVTQVEETKQMTKMVPILITTL 932
Query: 233 ---TMSINIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPL 289
TM I F + Q +LDR + NFE+P S ILI+++L I IYDRV +P+
Sbjct: 933 IPCTMLIQAHTLF-IKQGTTLDRSM--GPNFEIPPAGLSAILIISMLTSIPIYDRVFVPV 1103
Query: 290 ASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAM 349
+ P I+ +R+G GL L ++ A + E R + A +E +L H ++ +S
Sbjct: 1104IRRYTKNPRGITMLQRLGAGLLMYVLIMVIAWLTERKRLRVA-REKHLLGQHDIIPLSIF 1280
Query: 350 WLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIES 409
L PQ L G+AE F I + +F+Y + P+ M S+ S S ++ +G +SS +LS +
Sbjct: 1281ILIPQYALTGVAENFAEIAKMDFFYYQAPEGMKSLGISYSTTSVALGCFLSSFLLSTVAD 1460
Query: 410 TTPSGGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAYGPTVD 459
T ++GW+ DN+N D YY +V ++ LN L +LV + + VD
Sbjct: 1461ITKKHSHQGWILDNLNISRLDYYYAFMVILSFLNFLCFLVTAKFFDYNVD 1610
>AV411896
Length = 323
Score = 166 bits (420), Expect = 9e-42
Identities = 81/107 (75%), Positives = 93/107 (86%)
Frame = +1
Query: 114 LMLISTVLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKTSPEFYHQQKDSE 173
LML+STV F LASPLY+K K +TSL TGFAQV+VAA+KNRKL LPPK S E YH++KDS+
Sbjct: 1 LMLLSTVFFLLASPLYIKNKIQTSLITGFAQVTVAAFKNRKLSLPPKNSAELYHRRKDSD 180
Query: 174 LVVPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKA 220
LV+PTDKLRFLNKACVIKD +QDI SDGSA + W LCT+DQVEELKA
Sbjct: 181 LVIPTDKLRFLNKACVIKDPKQDITSDGSASDPWRLCTIDQVEELKA 321
>TC14955 UP|O22305 (O22305) Peptide transporter, complete
Length = 2300
Score = 138 bits (347), Expect = 3e-33
Identities = 100/380 (26%), Positives = 177/380 (46%), Gaps = 8/380 (2%)
Frame = +1
Query: 106 VGFGVPAILMLISTVLFFLASPLYV-KIKQKTSLFTGFAQVSVAAYKNRKLPLP--PKTS 162
+ + + AI L+S+++FF SP+Y K +Q S F +V + A++NRKL LP P
Sbjct: 667 LAYSLSAIGFLLSSIIFFWGSPVYRHKSRQARSPSMNFIRVPLVAFRNRKLQLPCNPSEL 846
Query: 163 PEF---YHQQKDSELVVPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELK 219
EF Y+ + + T FL++A I++ D+++ CTV QVE K
Sbjct: 847 HEFQLNYYISSGARKIHHTSHFSFLDRAA-IRESNTDLSNPP--------CTVTQVEGTK 999
Query: 220 AIIKVIPLWSTAI--TMSINIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILI 277
++ + +W + T + + + Q ++DR + F +PA S +++ LI
Sbjct: 1000 LVLGMFQIWLLMLIPTNCWALESTIFVRQGTTMDRTL--GPKFRLPAASLWCFIVLTTLI 1173
Query: 278 WIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYL 337
+ IYD IP + G I +R+GIG+ + + ET +R IK+ ++
Sbjct: 1174 CLPIYDHYFIPFMRRRTGNHRGIKLLQRVGIGMAIQVIAMAVTYAVET-QRMSVIKKHHI 1350
Query: 338 NDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGN 397
+ MS WL PQ + G++ F G EF+Y + P+ M + +L + G+
Sbjct: 1351 VGPEETVPMSIFWLLPQNIILGVSNAFLATGMLEFFYDQSPEEMKGLGTTLCTSCVAAGS 1530
Query: 398 LVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAYGPT 457
+++ ++++I+ W+ +N+N H D YY + I+ALN +L W
Sbjct: 1531 YINTFLVTMIDKL-------NWIGNNLNDSHLDYYYAFLFVISALNFGVFL---WVSSGY 1680
Query: 458 VDEVSNVSKENGSKVEESTE 477
+ + N S +E S E
Sbjct: 1681 IYKKENTSTTEVHDIEMSAE 1740
Score = 55.5 bits (132), Expect = 2e-08
Identities = 40/123 (32%), Positives = 67/123 (53%), Gaps = 1/123 (0%)
Frame = +3
Query: 3 LLWLTAMIPSARPPACNHPSEGCESATPGQLAMLFSALILIAIGNGGISCSLA-FGADQV 61
LL LT + S RP AC + C A+ Q+A+ +++L IA+G+G + +++ FGADQ
Sbjct: 366 LLVLTTTLKSLRP-ACENGI--CREASNLQVALFYTSLYTIAVGSGAVKPNMSTFGADQF 536
Query: 62 NRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLISTVL 121
+ + + + FF+W+ ++A +VYIQ+ G GFG+ I ST L
Sbjct: 537 DDFRHEEKEQKVS-FFNWWAFNGACGSLMATLFVVYIQEKNG--XGFGLQFICYWFSTFL 707
Query: 122 FFL 124
++L
Sbjct: 708 YYL 716
>TC18768
Length = 738
Score = 107 bits (268), Expect(2) = 9e-29
Identities = 63/196 (32%), Positives = 105/196 (53%), Gaps = 2/196 (1%)
Frame = +2
Query: 161 TSPEFYHQQKDSELVVPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKA 220
++P+ H +++ V TDK RFL+KAC+ + A + + W LC+V QVE++K
Sbjct: 188 SNPQMLHGDQNNVGQVHTDKFRFLDKACIRVEE----AGSNTKKSSWRLCSVGQVEQVKI 355
Query: 221 IIKVIPLWSTAITMSINIGG--SFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIW 278
++ VIP++S I + + +F + Q ++D H+ S F +P S I + ++I
Sbjct: 356 LLSVIPIFSCTIVFNTILAQLQTFSVQQGSAMDTHLTKS--FHIPPASLQSIPYILLIIV 529
Query: 279 IIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLN 338
+ +YD +P A +I G ISP +R+G GLF +++AA+ E RR EA+ N
Sbjct: 530 VPLYDTFFVPFARRITGHESGISPLRRIGFGLFLATFSMVSAALLEKKRRDEALNH---N 700
Query: 339 DTHGVLKMSAMWLAPQ 354
T +S W+ PQ
Sbjct: 701 KT-----LSIFWITPQ 733
Score = 36.2 bits (82), Expect(2) = 9e-29
Identities = 24/68 (35%), Positives = 33/68 (48%), Gaps = 3/68 (4%)
Frame = +1
Query: 98 IQDHLGWKVGFGVPAILMLISTVLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPL 157
IQ H G VGFG+ A +M + + + Y + S+ T AQV VAA+ +K L
Sbjct: 4 IQTHSGMDVGFGISAAVMAMGLISLISGTFYYRNKPPQGSILTPIAQVLVAAFF*KKAYL 183
Query: 158 ---PPKTS 162
PP S
Sbjct: 184 SI*PPNAS 207
>TC16709 weakly similar to PIR|T47573|T47573 peptide transport-like protein
- Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(41%)
Length = 712
Score = 120 bits (302), Expect = 4e-28
Identities = 69/235 (29%), Positives = 127/235 (53%), Gaps = 9/235 (3%)
Frame = +1
Query: 111 PAILMLISTVLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKTSPEFYHQQK 170
PA+ M ++ V FF + LY K S T QV VA+ + + P S ++
Sbjct: 1 PAVAMAVAVVSFFSGTRLYRNQKPGGSPLTRMCQVVVASMRKCGVQAPDDKS--LLYEIA 174
Query: 171 DSELVVP-------TDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIK 223
D+E + T++L F +KA V+ + +++ W LCT QVEELK+I++
Sbjct: 175 DTESAIKGSRKLDHTNELSFFDKAAVVVQS----SDQKDSVDPWRLCTETQVEELKSILR 342
Query: 224 VIPLWSTAITMSINIG--GSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIII 281
++P+W+T I + G + +LQ ++++ H+ +SNF++P + S+ ++++ W+ +
Sbjct: 343 MLPVWATGIIFATVYGQMSTLFVLQGQTMNTHV-GNSNFKIPPAALSIFDTLSVIFWVPV 519
Query: 282 YDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGY 336
YD +++P+A K G ++ +RMGIGLF + ++ AAI E +R + + Y
Sbjct: 520 YDWIIVPIARKFSGHKNGLTQLQRMGIGLFISIFAMVAAAILEVIRLRMVRRHNY 684
>BU494138
Length = 518
Score = 114 bits (286), Expect = 3e-26
Identities = 64/168 (38%), Positives = 95/168 (56%), Gaps = 2/168 (1%)
Frame = +2
Query: 205 NRWSLCTVDQVEELKAIIKVIPLWSTAIT--MSINIGGSFGLLQAKSLDRHIISSSNFEV 262
N + ++ QVEE+K + ++ P+W+ I ++ G+F + QA +DRHI S F++
Sbjct: 2 NSARVVSIQQVEEIKCLARIFPIWAAGILGFTAMAQQGTFTVSQAMKMDRHI--GSKFQI 175
Query: 263 PAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAI 322
PAGS VI + I +W+ YDR +P +I I+ +R+GIG+ F+ L +I A +
Sbjct: 176 PAGSLGVISFITIGLWVPFYDRFFVPAVRRITKHEGGITLLQRIGIGMVFSVLSMIVAGL 355
Query: 323 FETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQN 370
E VRR A + G+ MS MWLAPQL L G+ E FN IG N
Sbjct: 356 VEKVRRGVANSN---PNPLGIAPMSVMWLAPQLVLMGLCEAFNAIGTN 490
>TC18700 similar to UP|Q9LYR6 (Q9LYR6) Peptide transporter-like protein,
partial (18%)
Length = 615
Score = 99.8 bits (247), Expect = 1e-21
Identities = 48/146 (32%), Positives = 83/146 (55%), Gaps = 1/146 (0%)
Frame = +1
Query: 332 IKEGY-LNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSG 390
I+ G+ N + +SA WL Q CL G+AE+F ++G EF Y+E P +M S+ ++ +
Sbjct: 1 IEHGFEFNFLSAMPNLSAYWLLIQYCLIGVAEVFCIVGLLEFLYEEAPDAMKSIGSAYAA 180
Query: 391 LAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVC 450
LA G+G ++++ SII+S T G E W++ NIN G FD +YW++ ++ +N ++
Sbjct: 181 LAGGLGCFAATIINSIIKSLTGKEGKESWLAQNINTGRFDYFYWILTALSLVNFCIFIYS 360
Query: 451 SWAYGPTVDEVSNVSKENGSKVEEST 476
+ Y +V + K + + ST
Sbjct: 361 AHRYKYRTQQVYEMEKHDVANNVSST 438
>CN824992
Length = 708
Score = 97.8 bits (242), Expect = 4e-21
Identities = 69/229 (30%), Positives = 112/229 (48%), Gaps = 5/229 (2%)
Frame = +2
Query: 1 MSLLWLTAMIPSARPPACNHPSEGCESATPGQLAMLFSALILIAIGNGGISCSLA-FGAD 59
++ L LT + P C C S + Q A+ + ++ LIA+GNGG ++A FGAD
Sbjct: 32 LASLSLTTYLFLLEPNGCGDKELPCTSHSSYQTALFYVSIYLIALGNGGYQPNIATFGAD 211
Query: 60 QVNRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLIST 119
Q + +D P + FFS++Y + + + T + Y +D W +GF A ++
Sbjct: 212 QFDEED-PKEQHSKISFFSYFYLALNLGSLFSNTILNYFEDDGLWTLGFWASAGSAFLAL 388
Query: 120 VLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKTSPEFYHQQKDS----ELV 175
VLF +P Y K + F QV VAA + K+ + P + + ++ S +
Sbjct: 389 VLFLCGTPWYRYFKPGGNPVPRFCQVFVAAMRKWKVKVSPGEEDQLHEVEECSTNGGRKM 568
Query: 176 VPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKV 224
T+ RFL+KA +I E ++ S W L TV QVEE+K I+++
Sbjct: 569 FHTEGFRFLDKAALITSQESKQENECSL---WHLSTVTQVEEVKCILRL 706
>TC19180 similar to PIR|T10255|T10255 nitrite transport protein, chloroplast
- cucumber {Cucumis sativus;} , partial (12%)
Length = 408
Score = 91.3 bits (225), Expect = 4e-19
Identities = 53/126 (42%), Positives = 75/126 (59%), Gaps = 2/126 (1%)
Frame = +1
Query: 205 NRWSLCTVDQVEELKAIIKVIPLWSTAITMSINIG--GSFGLLQAKSLDRHIISSSNFEV 262
N+W L TV +VEELK+II++ P+W++ I + G+F L QAK++DRHI S F++
Sbjct: 34 NKWRLNTVHRVEELKSIIRMGPIWASGILLITAYAQQGTFSLQQAKTMDRHITKS--FQI 207
Query: 263 PAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAI 322
PAGS SV I+ +L +YDRVLI +A + G IS RMGIG + + A
Sbjct: 208 PAGSMSVFTIITMLTTTALYDRVLIRVARRFTGLDRGISFLHRMGIGFVISTIATFVAGF 387
Query: 323 FETVRR 328
E R+
Sbjct: 388 VEMKRK 405
>CB826936
Length = 596
Score = 89.0 bits (219), Expect = 2e-18
Identities = 51/166 (30%), Positives = 83/166 (49%)
Frame = +3
Query: 300 ISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAG 359
I+ +R+G GL + ++TA + E +R +E L H + ++ L PQ L G
Sbjct: 36 IAMLQRLGAGLVMYIITMVTAWLSER-KRLSVAREYNLLGQHDKIPLTIFILLPQFALTG 212
Query: 360 IAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGW 419
+A F I +F+Y + P+ M S+ S S + +G SS +LS + T G++GW
Sbjct: 213 VANNFVDIAMLDFFYDQAPEGMKSLGISYSTTSTALGCFFSSFLLSTVADLTKKHGHKGW 392
Query: 420 VSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAYGPTVDEVSNVS 465
V DN+N H D YY + ++ N L +LV + + VD + S
Sbjct: 393 VLDNLNVSHLDYYYAFMAILSVANFLCFLVAAKLFAYNVDVIHKKS 530
>AV409553
Length = 433
Score = 87.4 bits (215), Expect = 5e-18
Identities = 45/130 (34%), Positives = 77/130 (58%), Gaps = 1/130 (0%)
Frame = +2
Query: 1 MSLLWLTAMIPSARPPACNHPSEGCESATPGQLAMLFSALILIAIGNGGISCSLA-FGAD 59
M LL + + S +P N C A+ Q+ ++AL AIG GG +++ FGAD
Sbjct: 38 MILLTVAVSLKSLKPTCTNGI---CNKASTSQIVFFYTALYTTAIGAGGTKPNISTFGAD 208
Query: 60 QVNRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLIST 119
Q + NP+ ++ FF+W+ + + +IA G+VYIQ++LGW +G+G+P +L+S
Sbjct: 209 QFD-DFNPHEKQTKASFFNWWMFTSFLGALIATLGLVYIQENLGWGLGYGIPTSGLLLSL 385
Query: 120 VLFFLASPLY 129
V+F++ +P+Y
Sbjct: 386 VIFYVGTPMY 415
>TC8505 similar to UP|Q8LG02 (Q8LG02) Nitrate transporter, partial (23%)
Length = 1495
Score = 65.1 bits (157), Expect(2) = 2e-17
Identities = 29/86 (33%), Positives = 52/86 (59%)
Frame = +3
Query: 364 FNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDN 423
F +GQ EF+ +E P+ M S++ L A+ +G VSSL++SI++ + + W+ N
Sbjct: 249 FAYVGQLEFFIREAPERMKSMSTGLFLAALSMGYFVSSLLVSIVDKLS----KKKWLKSN 416
Query: 424 INKGHFDKYYWVIVGINALNLLYYLV 449
+NKG D +YW++ + LN + ++V
Sbjct: 417 LNKGRLDYFYWLLAVLGVLNFILFIV 494
Score = 40.8 bits (94), Expect(2) = 2e-17
Identities = 22/56 (39%), Positives = 31/56 (55%)
Frame = +2
Query: 307 GIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAE 362
GIGL F+F+ ++ +AI E RR+ A+K+ +SA WL PQ L G E
Sbjct: 101 GIGLVFSFVAMMVSAIVEKERRENAVKKH--------TNISAFWLVPQFFLVGAGE 244
>CB828533
Length = 557
Score = 83.2 bits (204), Expect = 1e-16
Identities = 48/133 (36%), Positives = 73/133 (54%), Gaps = 4/133 (3%)
Frame = +1
Query: 1 MSLLWLTAMIPSARPPACN---HPSEGCESATPGQLAMLFSALILIAIGNGGISCSLA-F 56
M LL IPS + P C+ C A+ Q+A+L AL IA+G GGI S++ F
Sbjct: 106 MCLLTGATTIPSMKAPPCSSVQRQHHECIEASGKQVALLLVALYTIAVGAGGIKSSVSGF 285
Query: 57 GADQVNRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILML 116
G+DQ + D P + + FF+ +Y F + + ++ +VY+QD++G GFG+PA +ML
Sbjct: 286 GSDQFDTTD-PREEKKMVFFFNRFYFFVSTGSLFSVLVLVYVQDNIGRGWGFGIPAGIML 462
Query: 117 ISTVLFFLASPLY 129
+S PLY
Sbjct: 463 VSLAFLLYGKPLY 501
>BG662103
Length = 387
Score = 82.4 bits (202), Expect = 2e-16
Identities = 39/108 (36%), Positives = 68/108 (62%), Gaps = 1/108 (0%)
Frame = +1
Query: 347 SAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSI 406
+++W+A Q G A++F + G EF++ E P M S+A SLS ++ +G +SS ++SI
Sbjct: 10 TSLWIAFQYLFLGSADLFTLAGLLEFFFSEAPIRMRSLATSLSWASLAIGYYLSSAIVSI 189
Query: 407 IESTTPSGGNEGWVSD-NINKGHFDKYYWVIVGINALNLLYYLVCSWA 453
+ S T G ++ W+S N+N H +++YW++ ++ LN L+YL WA
Sbjct: 190 VNSVTGKGSHKPWLSGANLNHYHLERFYWLMCLLSGLNFLHYLY--WA 327
>TC20040 similar to UP|Q9FM20 (Q9FM20) Nitrate transporter NTL1, partial
(20%)
Length = 578
Score = 79.0 bits (193), Expect = 2e-15
Identities = 36/110 (32%), Positives = 69/110 (62%), Gaps = 1/110 (0%)
Frame = +3
Query: 346 MSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLS 405
M+ +W+A Q G A++F + G EF++ E P SM S+A +LS ++ +G +S++++S
Sbjct: 3 MTFLWVAFQYLFLGSADLFTLAGMMEFFFTEAPWSMRSLATALSWASLSMGYFLSTVLVS 182
Query: 406 IIESTTPSGGNEGW-VSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAY 454
II T + G+ W + N+N H +++YW++ ++ LN ++YL + +Y
Sbjct: 183 IINKVTGAFGHTPWLLGSNLNHYHLERFYWLMCVLSGLNFIHYLFWANSY 332
>BP076619
Length = 432
Score = 75.9 bits (185), Expect = 2e-14
Identities = 29/98 (29%), Positives = 59/98 (59%)
Frame = -3
Query: 357 LAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGN 416
L G++E+F ++G EF+Y + P + S+ +L GVG+ +S ++S+IE+ T G
Sbjct: 427 LFGVSEVFTLVGLQEFFYDQVPNELRSLGLALYLSIFGVGSFLSGFLISVIETVTGKDGP 248
Query: 417 EGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAY 454
+ W ++N++K H D +YW++ G + + ++ + +Y
Sbjct: 247 DRWFANNLHKAHIDYFYWLLAGFSVVGFAMFMCFAKSY 134
>BP071089
Length = 393
Score = 72.0 bits (175), Expect = 2e-13
Identities = 31/75 (41%), Positives = 49/75 (65%)
Frame = -3
Query: 374 YKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYY 433
Y E P +M S+ ++LS +GN VS+L++ I+ T S G+ GW+ DN+N+GH D +Y
Sbjct: 391 YGEAPDAMRSLCSALSLTTNALGNYVSTLLVIIVTKVTTSSGSLGWIPDNMNRGHLDYFY 212
Query: 434 WVIVGINALNLLYYL 448
W++ ++ LN L YL
Sbjct: 211 WLLTVLSLLNFLVYL 167
>AV768501
Length = 442
Score = 67.8 bits (164), Expect = 4e-12
Identities = 28/88 (31%), Positives = 56/88 (62%)
Frame = -1
Query: 367 IGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINK 426
IGQ +F+ +E PK M +++ L + +G SS++++++ T G +GW++DN+N+
Sbjct: 442 IGQLDFFLRECPKGMKTMSPGLFLSTLSLGFFFSSMLVTLVHKMT--GPRQGWLADNLNQ 269
Query: 427 GHFDKYYWVIVGINALNLLYYLVCSWAY 454
G D +YW++ ++ +NL+ +L C+ Y
Sbjct: 268 GKLDDFYWLLAVLSGVNLVVFLFCAKWY 185
>AV768616
Length = 396
Score = 67.4 bits (163), Expect = 5e-12
Identities = 27/80 (33%), Positives = 52/80 (64%)
Frame = -1
Query: 375 KEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYW 434
++ P +M S++++LS L + +G +SSL+++I+ + G GW+ DN+N GH D ++W
Sbjct: 396 EQAPDAMRSLSSALSLLTVSLGQYLSSLLVTIVTKISTKNGGSGWIPDNLNYGHVDYFFW 217
Query: 435 VIVGINALNLLYYLVCSWAY 454
++ ++ LNL+ YL + Y
Sbjct: 216 LLAVLSVLNLIVYLPVAKLY 157
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,599,294
Number of Sequences: 28460
Number of extensions: 135421
Number of successful extensions: 722
Number of sequences better than 10.0: 64
Number of HSP's better than 10.0 without gapping: 698
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 703
length of query: 526
length of database: 4,897,600
effective HSP length: 94
effective length of query: 432
effective length of database: 2,222,360
effective search space: 960059520
effective search space used: 960059520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC149489.11


