

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149307.14 - phase: 0
(629 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
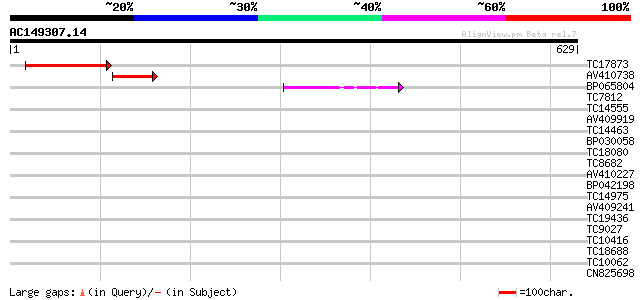
Score E
Sequences producing significant alignments: (bits) Value
TC17873 similar to UP|Q8RX56 (Q8RX56) AT5g06970/MOJ9_14, partial... 185 2e-47
AV410738 92 3e-19
BP065804 86 1e-17
TC7812 UP|Q9LKJ6 (Q9LKJ6) Water-selective transport intrinsic me... 39 0.003
TC14555 similar to UP|Q43558 (Q43558) Proline rich protein precu... 38 0.006
AV409919 37 0.007
TC14463 similar to UP|Q9SLF1 (Q9SLF1) Nodulin-like protein (At2g... 37 0.010
BP030058 37 0.010
TC18080 similar to GB|AAL90935.1|19699138|AY090274 AT3g18790/MVE... 37 0.012
TC8682 similar to UP|HPPD_SOLSC (Q9ARF9) 4-hydroxyphenylpyruvate... 37 0.012
AV410227 36 0.021
BP042198 35 0.028
TC14975 homologue to UP|Q8LG37 (Q8LG37) AtPH1-like protein, part... 31 0.032
AV409241 35 0.036
TC19436 similar to UP|Q93Y06 (Q93Y06) Receptor protein kinase-li... 34 0.062
TC9027 weakly similar to UP|AAQ82447 (AAQ82447) SufB, partial (51%) 34 0.062
TC10416 weakly similar to UP|O04108 (O04108) MYB factor, partial... 34 0.081
TC18688 weakly similar to UP|Q9LP77 (Q9LP77) T1N15.9, partial (13%) 34 0.081
TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein... 33 0.11
CN825698 33 0.11
>TC17873 similar to UP|Q8RX56 (Q8RX56) AT5g06970/MOJ9_14, partial (9%)
Length = 602
Score = 185 bits (469), Expect = 2e-47
Identities = 91/96 (94%), Positives = 94/96 (97%)
Frame = +2
Query: 18 ENAIDLLQRYRRDRRVLLDFILSGSLIKKVVMPPGAVTLDDVDLDQVSIDYVLNCAKKSE 77
ENAIDLLQRYRRDRRVLLDF+LSGSLIKKVVMPPGAVTLDDVDLDQVS+DYV+NCAKKS
Sbjct: 314 ENAIDLLQRYRRDRRVLLDFMLSGSLIKKVVMPPGAVTLDDVDLDQVSVDYVVNCAKKSV 493
Query: 78 MLELSEAIRDYHDHTGLPQMSDTGSVGEFYLVTDPE 113
LELSEAIRDYHDHTGLPQMSDTGSVGEFYLVTDPE
Sbjct: 494 TLELSEAIRDYHDHTGLPQMSDTGSVGEFYLVTDPE 601
>AV410738
Length = 153
Score = 91.7 bits (226), Expect = 3e-19
Identities = 41/50 (82%), Positives = 46/50 (92%)
Frame = +2
Query: 115 SGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVASNISRSESLYSAQER 164
SGSPPKRAPPPVPISA PP+AVSTPPPAYPTSP+AS++SRSES S QE+
Sbjct: 2 SGSPPKRAPPPVPISAAPPVAVSTPPPAYPTSPIASSVSRSESFNSTQEK 151
>BP065804
Length = 492
Score = 86.3 bits (212), Expect = 1e-17
Identities = 46/134 (34%), Positives = 77/134 (57%)
Frame = +2
Query: 304 GKRMDTLLVPLELLCCVARTEFSDKKAFIRWQKRQLKVLEEGLVNHPVVGFGESGRKTNE 363
G++ +T+++PLELL + +EFS+ + WQKRQLK+LE GL+ HP + ++
Sbjct: 104 GRQAETIILPLELLRHLKPSEFSNPHEYHLWQKRQLKILEAGLLXHPSIPVEKNNTFAMN 283
Query: 364 MRILLAKIEESEFLPSSSGELQRTECLRSLREIAIPLAERPARGDLTGEICHWADGYQFN 423
+R + I +E P + + +E +R+ + L+ R G + +CHWA+GY N
Sbjct: 284 LRDI---INSAELQPLDTS--KTSETMRTFSNAVVSLSMRSPNG-IPTNVCHWANGYPVN 445
Query: 424 VRLYEKLLLSVFDM 437
+ LY LL S+FD+
Sbjct: 446 IHLYISLLQSIFDL 487
>TC7812 UP|Q9LKJ6 (Q9LKJ6) Water-selective transport intrinsic membrane
protein 1, complete
Length = 1149
Score = 38.5 bits (88), Expect = 0.003
Identities = 25/65 (38%), Positives = 31/65 (47%), Gaps = 15/65 (23%)
Frame = +1
Query: 112 PESSGSP-----------PKRAPPPVPISAVPPIAV----STPPPAYPTSPVASNISRSE 156
P SS SP P APPP P S+ PP S+P P+ PTSP A++ S
Sbjct: 163 PSSSSSPAPVPESPTTNSPTTAPPPPPASSPPPSPTHSPSSSPSPSAPTSPAATSTPPSP 342
Query: 157 SLYSA 161
S S+
Sbjct: 343 SASSS 357
Score = 28.1 bits (61), Expect = 4.4
Identities = 17/47 (36%), Positives = 23/47 (48%)
Frame = +1
Query: 104 GEFYLVTDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVAS 150
G+ +L + P SS S +P PVP S + PPP +SP S
Sbjct: 136 GQPWLSSSPPSSSS----SPAPVPESPTTNSPTTAPPPPPASSPPPS 264
>TC14555 similar to UP|Q43558 (Q43558) Proline rich protein precursor,
partial (43%)
Length = 1038
Score = 37.7 bits (86), Expect = 0.006
Identities = 18/33 (54%), Positives = 19/33 (57%)
Frame = +2
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAYP 144
P S PP +PPPV S PP STPPPA P
Sbjct: 332 PAQSSPPPVSSPPPVQ-STPPPAPASTPPPASP 427
Score = 36.6 bits (83), Expect = 0.012
Identities = 18/41 (43%), Positives = 22/41 (52%), Gaps = 2/41 (4%)
Frame = +2
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVS--TPPPAYPTSPVAS 150
P SS P + PPP P S PP + +PPPA P P A+
Sbjct: 353 PVSSPPPVQSTPPPAPASTPPPASPPPFSPPPATPPPPAAT 475
Score = 36.2 bits (82), Expect = 0.016
Identities = 17/40 (42%), Positives = 21/40 (52%), Gaps = 1/40 (2%)
Frame = +2
Query: 112 PESSGSPPKRAPPPV-PISAVPPIAVSTPPPAYPTSPVAS 150
P + +PP +PPP P A PP +TPPPA P S
Sbjct: 392 PAPASTPPPASPPPFSPPPATPPPPAATPPPALTPVPATS 511
Score = 35.8 bits (81), Expect = 0.021
Identities = 18/41 (43%), Positives = 23/41 (55%), Gaps = 5/41 (12%)
Frame = +2
Query: 112 PESSGSPPKRAPPPV-----PISAVPPIAVSTPPPAYPTSP 147
P S PP PPP P+S+ PP+ STPPPA ++P
Sbjct: 293 PVQSSPPPASTPPPAQSSPPPVSSPPPVQ-STPPPAPASTP 412
Score = 35.0 bits (79), Expect = 0.036
Identities = 17/40 (42%), Positives = 22/40 (54%)
Frame = +2
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVASN 151
P + SPP P P+ + PP A STPPPA + P S+
Sbjct: 248 PAAQSSPPPAQSSPPPVQSSPPPA-STPPPAQSSPPPVSS 364
Score = 34.7 bits (78), Expect = 0.047
Identities = 15/33 (45%), Positives = 18/33 (54%)
Frame = +2
Query: 115 SGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSP 147
S SP PP P + PP A S+PPPA + P
Sbjct: 194 SNSPTTSPAPPTPTTPQPPAAQSSPPPAQSSPP 292
Score = 34.7 bits (78), Expect = 0.047
Identities = 18/42 (42%), Positives = 24/42 (56%), Gaps = 4/42 (9%)
Frame = +2
Query: 113 ESSGSPPKRAPPPV----PISAVPPIAVSTPPPAYPTSPVAS 150
+SS P + +PPPV P ++ PP A S+PPP PV S
Sbjct: 257 QSSPPPAQSSPPPVQSSPPPASTPPPAQSSPPPVSSPPPVQS 382
Score = 32.7 bits (73), Expect = 0.18
Identities = 17/38 (44%), Positives = 20/38 (51%)
Frame = +2
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVA 149
P SPP + PP S+ PP VS+PPP T P A
Sbjct: 290 PPVQSSPPPASTPPPAQSSPPP--VSSPPPVQSTPPPA 397
Score = 32.3 bits (72), Expect = 0.24
Identities = 20/57 (35%), Positives = 28/57 (49%), Gaps = 1/57 (1%)
Frame = +2
Query: 102 SVGEFYLVTDPESSGSPPKRAPPPVPIS-AVPPIAVSTPPPAYPTSPVASNISRSES 157
SVG P +S +PP P P + + PP A S+PPP + P AS ++S
Sbjct: 173 SVGAQSPSNSPTTSPAPPTPTTPQPPAAQSSPPPAQSSPPPVQSSPPPASTPPPAQS 343
>AV409919
Length = 423
Score = 37.4 bits (85), Expect = 0.007
Identities = 23/69 (33%), Positives = 30/69 (43%), Gaps = 2/69 (2%)
Frame = +3
Query: 88 YHDHTGLPQMSDTGSVGEFYLVTDPESSGSPPKRAPPPVPISAVPPI--AVSTPPPAYPT 145
Y + P + + + G T P S+ SPP R PP P S P A S+ PP T
Sbjct: 177 YFSPSSSPSSTSSSTAGARRFGTPPLSTSSPPPRPPPSSPSSPPPSTSSASSSAPPPTTT 356
Query: 146 SPVASNISR 154
SP + R
Sbjct: 357 SPTTRSWPR 383
Score = 28.1 bits (61), Expect = 4.4
Identities = 18/54 (33%), Positives = 24/54 (44%), Gaps = 4/54 (7%)
Frame = +3
Query: 108 LVTDPESSGSPPKRAPPPVPISAVPPIAVST----PPPAYPTSPVASNISRSES 157
L P SS S + PP++ S+ PPP+ P+SP S S S S
Sbjct: 174 LYFSPSSSPSSTSSSTAGARRFGTPPLSTSSPPPRPPPSSPSSPPPSTSSASSS 335
>TC14463 similar to UP|Q9SLF1 (Q9SLF1) Nodulin-like protein
(At2g16660/T24I21.7), partial (40%)
Length = 1122
Score = 37.0 bits (84), Expect = 0.010
Identities = 17/37 (45%), Positives = 20/37 (53%)
Frame = -3
Query: 111 DPESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSP 147
+P SS P APPP P + PP PPP+ P SP
Sbjct: 1093 NPHSSSPPISCAPPPPPAAPSPPPLPLPPPPSPPRSP 983
>BP030058
Length = 455
Score = 37.0 bits (84), Expect = 0.010
Identities = 20/45 (44%), Positives = 22/45 (48%), Gaps = 5/45 (11%)
Frame = -1
Query: 112 PESSGSPPKR-----APPPVPISAVPPIAVSTPPPAYPTSPVASN 151
P S SPP +PPP P S PP S PPP YP+ P N
Sbjct: 401 PPPSPSPPPPPLPCPSPPPPPPSPPPPPPPSPPPPKYPSCPQNCN 267
Score = 32.7 bits (73), Expect = 0.18
Identities = 14/35 (40%), Positives = 19/35 (54%)
Frame = -1
Query: 113 ESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSP 147
++ G PP PPP P PP+ +PPP P+ P
Sbjct: 425 DTCGDPPP--PPPSPSPPPPPLPCPSPPPPPPSPP 327
Score = 29.3 bits (64), Expect = 2.0
Identities = 14/30 (46%), Positives = 14/30 (46%)
Frame = -1
Query: 118 PPKRAPPPVPISAVPPIAVSTPPPAYPTSP 147
PP PPP P S PP S P P SP
Sbjct: 344 PPPSPPPPPPPSPPPPKYPSCPQNCNPLSP 255
>TC18080 similar to GB|AAL90935.1|19699138|AY090274 AT3g18790/MVE11_17
{Arabidopsis thaliana;}, partial (24%)
Length = 585
Score = 36.6 bits (83), Expect = 0.012
Identities = 17/42 (40%), Positives = 24/42 (56%), Gaps = 2/42 (4%)
Frame = -1
Query: 110 TDPESSGSPPKRAPPPVPISAVPPIAVSTPPP--AYPTSPVA 149
+ P S SPP+ PPP P+ + P PPP ++P+SP A
Sbjct: 141 SSPYPSSSPPQPHPPPPPLESPSPQPTPQPPPRFSHPSSPPA 16
>TC8682 similar to UP|HPPD_SOLSC (Q9ARF9) 4-hydroxyphenylpyruvate
dioxygenase (4HPPD) (HPD) (HPPDase) , partial (29%)
Length = 576
Score = 36.6 bits (83), Expect = 0.012
Identities = 16/39 (41%), Positives = 21/39 (53%)
Frame = +1
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVAS 150
P + P PPP P+S PP + S PP A P++P S
Sbjct: 415 PHTLPKSPSPPPPPSPLSPPPPASPSPPPTASPSAPSPS 531
Score = 31.2 bits (69), Expect = 0.52
Identities = 19/50 (38%), Positives = 23/50 (46%), Gaps = 2/50 (4%)
Frame = +1
Query: 110 TDPESSGSPPKRAP--PPVPISAVPPIAVSTPPPAYPTSPVASNISRSES 157
T P+S PP +P PP P S PP S P+ SP S S +S
Sbjct: 421 TLPKSPSPPPPPSPLSPPPPASPSPPPTASPSAPSPSKSPTPSKPSPPQS 570
>AV410227
Length = 428
Score = 35.8 bits (81), Expect = 0.021
Identities = 18/48 (37%), Positives = 23/48 (47%), Gaps = 9/48 (18%)
Frame = +1
Query: 112 PESSGSPPKRAP---------PPVPISAVPPIAVSTPPPAYPTSPVAS 150
P S+ SPP+ P P P+S P TPPP+ P SP +S
Sbjct: 124 PSSNASPPRPPPSTASSSATSPTSPLSTSPTTPTPTPPPSSPPSPASS 267
Score = 32.0 bits (71), Expect = 0.31
Identities = 16/41 (39%), Positives = 18/41 (43%)
Frame = +1
Query: 110 TDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVAS 150
T P S S P P P + PP S+PPP T P S
Sbjct: 181 TSPTSPLSTSPTTPTPTPPPSSPPSPASSPPPPSMTPPAPS 303
Score = 31.6 bits (70), Expect = 0.40
Identities = 20/38 (52%), Positives = 21/38 (54%)
Frame = +1
Query: 110 TDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSP 147
T P SS PP A P P S PP A STPPP+ SP
Sbjct: 229 TPPPSS--PPSPASSPPPPSMTPP-APSTPPPSSA*SP 333
Score = 30.0 bits (66), Expect = 1.2
Identities = 13/37 (35%), Positives = 20/37 (53%)
Frame = +1
Query: 108 LVTDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAYP 144
L T P + P + PP P S+ PP +++ P P+ P
Sbjct: 199 LSTSPTTPTPTPPPSSPPSPASSPPPPSMTPPAPSTP 309
Score = 27.3 bits (59), Expect = 7.6
Identities = 12/28 (42%), Positives = 14/28 (49%)
Frame = +1
Query: 114 SSGSPPKRAPPPVPISAVPPIAVSTPPP 141
+S PP PP P + P A S PPP
Sbjct: 259 ASSPPPPSMTPPAPSTPPPSSA*SPPPP 342
Score = 27.3 bits (59), Expect = 7.6
Identities = 15/38 (39%), Positives = 19/38 (49%), Gaps = 5/38 (13%)
Frame = +1
Query: 112 PESSGSPPKR-----APPPVPISAVPPIAVSTPPPAYP 144
P S+ SPP +PP V + PP A S PP +P
Sbjct: 313 PSSA*SPPPPPS*AGSPPAVEPLSAPPCANSPSPPRFP 426
>BP042198
Length = 481
Score = 35.4 bits (80), Expect = 0.028
Identities = 18/39 (46%), Positives = 22/39 (56%)
Frame = -3
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVAS 150
P SS S +PPPVP A PP TP + +SPVA+
Sbjct: 467 PASSPSNNVSSPPPVPSPATPPPTAETPAVSPVSSPVAA 351
>TC14975 homologue to UP|Q8LG37 (Q8LG37) AtPH1-like protein, partial (86%)
Length = 892
Score = 30.8 bits (68), Expect(2) = 0.032
Identities = 18/55 (32%), Positives = 26/55 (46%), Gaps = 1/55 (1%)
Frame = +3
Query: 102 SVGEFYLVTDPESSGSPPKRAPPPVPISAV-PPIAVSTPPPAYPTSPVASNISRS 155
+ G F + S +PP RA P S+ PP++ S P T+P SN R+
Sbjct: 228 AAGSFSNKENSSGSKTPPSRASPSHAASSPSPPVSPSRAPKTSSTNPTPSNSPRA 392
Score = 23.1 bits (48), Expect(2) = 0.032
Identities = 20/68 (29%), Positives = 34/68 (49%), Gaps = 10/68 (14%)
Frame = +2
Query: 147 PVASNIS-RSESLY----SAQERELTVDDI-----EDFEDDDDTSMVEGLRAKRTLNDAS 196
P A +S RS+++Y S +E+E ++ I + D+ +V+ AKR N +S
Sbjct: 365 PYAFELSTRSDTMYFIADSDKEKEDWINSIGRSIVQHSRSVTDSEIVDYDSAKR*RNSSS 544
Query: 197 DLAVKLPP 204
L + PP
Sbjct: 545 PLTINTPP 568
>AV409241
Length = 282
Score = 35.0 bits (79), Expect = 0.036
Identities = 19/44 (43%), Positives = 22/44 (49%), Gaps = 1/44 (2%)
Frame = +2
Query: 115 SGSPPKRAPPPVPISAVPPIAVSTPP-PAYPTSPVASNISRSES 157
S SP K PPP P PP VS PP P+ P + S +S S
Sbjct: 83 SHSPTKTPPPPTPPPPPPPATVSPPPTPSPPAAATPSPLSAKPS 214
Score = 33.1 bits (74), Expect = 0.14
Identities = 15/37 (40%), Positives = 20/37 (53%)
Frame = +2
Query: 118 PPKRAPPPVPISAVPPIAVSTPPPAYPTSPVASNISR 154
PP PPP P + V P +PP A SP+++ SR
Sbjct: 107 PPPTPPPPPPPATVSPPPTPSPPAAATPSPLSAKPSR 217
>TC19436 similar to UP|Q93Y06 (Q93Y06) Receptor protein kinase-like protein,
partial (19%)
Length = 570
Score = 34.3 bits (77), Expect = 0.062
Identities = 17/52 (32%), Positives = 27/52 (51%)
Frame = +2
Query: 106 FYLVTDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPVASNISRSES 157
F +++ P SS S PPP P+S PP + T P YP++ + + S +
Sbjct: 38 FLIISKPHSSLSSSSLPPPP-PLSMTPPRSHQTQSPCYPSNAKQTKTTSSST 190
>TC9027 weakly similar to UP|AAQ82447 (AAQ82447) SufB, partial (51%)
Length = 1016
Score = 34.3 bits (77), Expect = 0.062
Identities = 14/35 (40%), Positives = 19/35 (54%)
Frame = +3
Query: 108 LVTDPESSGSPPKRAPPPVPISAVPPIAVSTPPPA 142
L + S PP+ PPP P + +PP TPPP+
Sbjct: 81 LTSSTHSRPQPPQTPPPPKPPNLLPPPPPQTPPPS 185
>TC10416 weakly similar to UP|O04108 (O04108) MYB factor, partial (41%)
Length = 1046
Score = 33.9 bits (76), Expect = 0.081
Identities = 32/96 (33%), Positives = 43/96 (44%), Gaps = 5/96 (5%)
Frame = +2
Query: 89 HDHTGLPQMSDTGSVGEFYLVTDPESSGSPPKRAPPPVPISAVPPIAVSTPPPAYPTSPV 148
H HT L + +V E VT+ E+ S K PVP S + + V TPPPA S
Sbjct: 410 HWHTTLKR-----TVHEQNTVTNEETKVSNSKNERDPVPKSGIASLLV-TPPPATSISGS 571
Query: 149 ASNIS--RSESLYSAQERELTVDDIED---FEDDDD 179
+S + S +S +LT + FEDD D
Sbjct: 572 IGALSPFSTSSEFSCTTSDLTTSSSSEMLVFEDDFD 679
>TC18688 weakly similar to UP|Q9LP77 (Q9LP77) T1N15.9, partial (13%)
Length = 514
Score = 33.9 bits (76), Expect = 0.081
Identities = 26/60 (43%), Positives = 32/60 (53%), Gaps = 12/60 (20%)
Frame = +3
Query: 110 TDPESS--GSPPKRAP--------PPVPISAVPPIAVSTPPPAY--PTSPVASNISRSES 157
T P SS SPP +P PP P SA+ ++VSTP PA+ PTSP A + S S
Sbjct: 243 TPPTSSRSASPPSPSPANSHMASSPPSPTSALS-VSVSTPSPAHSPPTSPPAPRSATSTS 419
>TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein 2, partial
(66%)
Length = 481
Score = 33.5 bits (75), Expect = 0.11
Identities = 16/37 (43%), Positives = 17/37 (45%), Gaps = 1/37 (2%)
Frame = -1
Query: 112 PESSGSPPKRAPPPVPISAV-PPIAVSTPPPAYPTSP 147
P PP R PPP P PP+ PPP YP P
Sbjct: 475 PPPPPRPPPRPPPPYPPPPP*PPLPPYPPPPPYPPPP 365
Score = 28.1 bits (61), Expect = 4.4
Identities = 13/30 (43%), Positives = 14/30 (46%)
Frame = -1
Query: 118 PPKRAPPPVPISAVPPIAVSTPPPAYPTSP 147
PP R PPP P PP PP P +P
Sbjct: 373 PPPRPPPP*PPPPYPPPP*PPPPRPPPPAP 284
Score = 28.1 bits (61), Expect = 4.4
Identities = 16/33 (48%), Positives = 17/33 (51%)
Frame = -1
Query: 118 PPKRAPPPVPISAVPPIAVSTPPPAYPTSPVAS 150
PP R PPP P PP V P A+P PV S
Sbjct: 313 PPPRPPPPAP----PPRRV--PCTAWPLGPVTS 233
Score = 28.1 bits (61), Expect = 4.4
Identities = 14/30 (46%), Positives = 14/30 (46%)
Frame = -1
Query: 118 PPKRAPPPVPISAVPPIAVSTPPPAYPTSP 147
PP PPP P PP PPP YP P
Sbjct: 403 PPYPPPPPYPPPPRPP--PP*PPPPYPPPP 320
>CN825698
Length = 704
Score = 33.5 bits (75), Expect = 0.11
Identities = 14/30 (46%), Positives = 16/30 (52%)
Frame = +1
Query: 112 PESSGSPPKRAPPPVPISAVPPIAVSTPPP 141
P S+ +PP PPP P S PP PPP
Sbjct: 115 PSSASAPPPPPPPPPPPSLPPPSDSDLPPP 204
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.136 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,323,600
Number of Sequences: 28460
Number of extensions: 149858
Number of successful extensions: 3443
Number of sequences better than 10.0: 333
Number of HSP's better than 10.0 without gapping: 2130
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2942
length of query: 629
length of database: 4,897,600
effective HSP length: 96
effective length of query: 533
effective length of database: 2,165,440
effective search space: 1154179520
effective search space used: 1154179520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC149307.14


