

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149305.9 - phase: 0 /pseudo
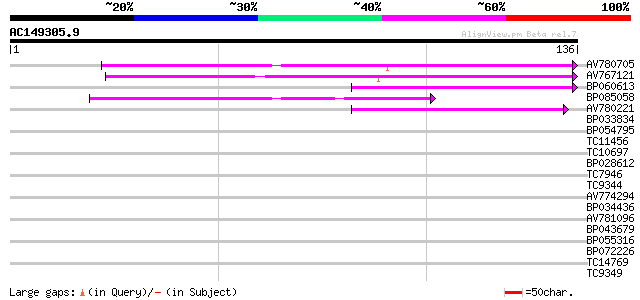
(136 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV780705 54 7e-09
AV767121 53 2e-08
BP060613 47 1e-06
BP085058 46 2e-06
AV780221 40 1e-04
BP033834 27 0.94
BP054795 26 2.7
TC11456 homologue to UP|Q9FI68 (Q9FI68) Arginine methyltransfera... 25 3.6
TC10697 25 3.6
BP028612 25 3.6
TC7946 homologue to UP|EF2_BETVU (O23755) Elongation factor 2 (E... 25 4.6
TC9344 similar to UP|AAQ84317 (AAQ84317) Fiber NTGP1-related pro... 25 6.1
AV774294 25 6.1
BP034436 24 7.9
AV781096 24 7.9
BP043679 24 7.9
BP055316 24 7.9
BP072226 24 7.9
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 24 7.9
TC9349 homologue to UP|Q8L3U4 (Q8L3U4) AT5g36790/f5h8_20, partia... 24 7.9
>AV780705
Length = 524
Score = 54.3 bits (129), Expect = 7e-09
Identities = 34/116 (29%), Positives = 56/116 (47%), Gaps = 2/116 (1%)
Frame = -2
Query: 23 KLASWKCRLLNKPSRVILVNSVLTSIPAYKMQIH*LAHYICVIDHLNKTVHSFI*K*STN 82
KL WK LLN+ + +L+ +++ +IP+Y M I C + L V F K
Sbjct: 409 KLEGWKETLLNQAGKEVLIKAIIQAIPSYAMTIVHFPKTFC--NSLYALVADFWWKSQGE 236
Query: 83 KGMNMVG--WDKITKAKKNGGLGVRVVRKQNIALLGKLVWNLLMPTNHLWSQILSA 136
+ G K +K+ GG+G R +R QN+A L + W +L LW +++ +
Sbjct: 235 GAVEYTGAVGTK*PLSKEKGGVGFRDLRTQNLAFLARQAWRVLTNPEALWVRVMKS 68
>AV767121
Length = 525
Score = 52.8 bits (125), Expect = 2e-08
Identities = 35/114 (30%), Positives = 59/114 (51%), Gaps = 1/114 (0%)
Frame = +2
Query: 24 LASWKCRLLNKPSRVILVNSVLTSIPAYKMQIH*LAHYICVIDHLNKTVHSFI*K*STNK 83
L+ WK + ++ R+ L+ SVLT++P + + L I V + + +F+ S N+
Sbjct: 2 LSKWKQKTVSFGGRIFLIQSVLTALPLFFLSFFKLP--IGVGKSCVRLMRNFLWGGSENE 175
Query: 84 GMNM-VGWDKITKAKKNGGLGVRVVRKQNIALLGKLVWNLLMPTNHLWSQILSA 136
V W + K K+ GGLG++ + N ALLGK W L + LW +++ A
Sbjct: 176 NKIA*VKWTDVCKPKELGGLGIKDLFTFNKALLGK*RWRYLTEPDSLWRRVIEA 337
>BP060613
Length = 378
Score = 47.0 bits (110), Expect = 1e-06
Identities = 23/54 (42%), Positives = 31/54 (56%)
Frame = +1
Query: 83 KGMNMVGWDKITKAKKNGGLGVRVVRKQNIALLGKLVWNLLMPTNHLWSQILSA 136
+G++ WD +T+ K GGLG + QN ALL K W +L + LW QIL A
Sbjct: 67 RGVHWRKWDLLTELKDEGGLGFKDFEIQNQALLAKQAWRILHNPDALWVQILKA 228
>BP085058
Length = 452
Score = 46.2 bits (108), Expect = 2e-06
Identities = 30/83 (36%), Positives = 45/83 (54%)
Frame = +2
Query: 20 LINKLASWKCRLLNKPSRVILVNSVLTSIPAYKMQIH*LAHYICVIDHLNKTVHSFI*K* 79
+I++LA K LLNKP+R+ L VL++I Y MQ++ L IC D + V F+ +
Sbjct: 203 VISRLAC*KAYLLNKPARLTLAKPVLSAISMYVMQLNWLPQSIC--DQICTVVRHFVWE- 373
Query: 80 STNKGMNMVGWDKITKAKKNGGL 102
G+ +V W+ I K GL
Sbjct: 374 -NVNGIPLVAWETIQSRKSMVGL 439
>AV780221
Length = 461
Score = 40.4 bits (93), Expect = 1e-04
Identities = 20/52 (38%), Positives = 29/52 (55%)
Frame = +3
Query: 83 KGMNMVGWDKITKAKKNGGLGVRVVRKQNIALLGKLVWNLLMPTNHLWSQIL 134
K + V W + + K GGLGV+ + N ALLGK W +L + LW ++L
Sbjct: 138 KKIAWVKWSTVCRPKDEGGLGVQNLGLFNKALLGKWRWRMLKERDGLWYKVL 293
>BP033834
Length = 534
Score = 27.3 bits (59), Expect = 0.94
Identities = 15/43 (34%), Positives = 22/43 (50%)
Frame = +1
Query: 81 TNKGMNMVGWDKITKAKKNGGLGVRVVRKQNIALLGKLVWNLL 123
+ G ++V W K T K GLG R RK + + LV++ L
Sbjct: 10 SRNG*HLVSWGKATHVKNY*GLGFREARKDHRKYVIHLVYHHL 138
>BP054795
Length = 516
Score = 25.8 bits (55), Expect = 2.7
Identities = 15/49 (30%), Positives = 27/49 (54%)
Frame = -2
Query: 3 TNHFNIIFKLISLVCGFLINKLASWKCRLLNKPSRVILVNSVLTSIPAY 51
TN F + KL+SL + L SW+ L ++PS+ ++ V + P++
Sbjct: 185 TNKFQLSRKLLSL-----LTNLLSWQSILCSEPSKNLVCLRVRPTPPSW 54
>TC11456 homologue to UP|Q9FI68 (Q9FI68) Arginine methyltransferase-like
protein, partial (10%)
Length = 669
Score = 25.4 bits (54), Expect = 3.6
Identities = 12/20 (60%), Positives = 14/20 (70%)
Frame = +2
Query: 29 CRLLNKPSRVILVNSVLTSI 48
C L+KP +ILVNS L SI
Sbjct: 401 CLYLDKPDNLILVNSSLHSI 460
>TC10697
Length = 487
Score = 25.4 bits (54), Expect = 3.6
Identities = 11/27 (40%), Positives = 17/27 (62%)
Frame = -1
Query: 110 QNIALLGKLVWNLLMPTNHLWSQILSA 136
QNI +L +WNL +P ++ +IL A
Sbjct: 118 QNIIMLSHKIWNLNLPFLYVIPKILKA 38
>BP028612
Length = 322
Score = 25.4 bits (54), Expect = 3.6
Identities = 12/20 (60%), Positives = 14/20 (70%)
Frame = -3
Query: 29 CRLLNKPSRVILVNSVLTSI 48
C L+KP +ILVNS L SI
Sbjct: 146 CLYLDKPDNLILVNSSLHSI 87
>TC7946 homologue to UP|EF2_BETVU (O23755) Elongation factor 2 (EF-2),
partial (34%)
Length = 1009
Score = 25.0 bits (53), Expect = 4.6
Identities = 14/25 (56%), Positives = 16/25 (64%), Gaps = 3/25 (12%)
Frame = +1
Query: 27 WKCRLLNKP---SRVILVNSVLTSI 48
WK RLL+KP S V L SV TS+
Sbjct: 511 WKSRLLSKPLVVSTVFLTRSVDTSL 585
>TC9344 similar to UP|AAQ84317 (AAQ84317) Fiber NTGP1-related protein,
complete
Length = 995
Score = 24.6 bits (52), Expect = 6.1
Identities = 10/21 (47%), Positives = 14/21 (66%)
Frame = -3
Query: 44 VLTSIPAYKMQIH*LAHYICV 64
+LTS+ AY M H*L + C+
Sbjct: 660 ILTSLKAYNMVQH*LVFFACL 598
>AV774294
Length = 498
Score = 24.6 bits (52), Expect = 6.1
Identities = 8/21 (38%), Positives = 16/21 (76%)
Frame = -2
Query: 12 LISLVCGFLINKLASWKCRLL 32
L+S +CG+L++ + + CRL+
Sbjct: 272 LLSCLCGYLLSASSRYHCRLV 210
>BP034436
Length = 417
Score = 24.3 bits (51), Expect = 7.9
Identities = 8/18 (44%), Positives = 14/18 (77%)
Frame = +1
Query: 4 NHFNIIFKLISLVCGFLI 21
+HF+II +I ++CG L+
Sbjct: 70 HHFSIIIIIIIIICGLLL 123
>AV781096
Length = 497
Score = 24.3 bits (51), Expect = 7.9
Identities = 11/21 (52%), Positives = 14/21 (66%)
Frame = +1
Query: 18 GFLINKLASWKCRLLNKPSRV 38
G L+NKL WK +L+ PS V
Sbjct: 373 GTLLNKLRLWKGFMLSAPSLV 435
>BP043679
Length = 501
Score = 24.3 bits (51), Expect = 7.9
Identities = 9/19 (47%), Positives = 15/19 (78%)
Frame = +3
Query: 4 NHFNIIFKLISLVCGFLIN 22
N FNII+ + +L+ G+L+N
Sbjct: 372 NSFNIIWSIFALL*GYLLN 428
>BP055316
Length = 198
Score = 24.3 bits (51), Expect = 7.9
Identities = 8/18 (44%), Positives = 13/18 (71%)
Frame = -1
Query: 12 LISLVCGFLINKLASWKC 29
L S++C F+ +K SW+C
Sbjct: 75 LPSILCSFIADKPISWEC 22
>BP072226
Length = 206
Score = 24.3 bits (51), Expect = 7.9
Identities = 8/15 (53%), Positives = 12/15 (79%)
Frame = +1
Query: 85 MNMVGWDKITKAKKN 99
MN++GW KIT ++N
Sbjct: 13 MNIIGW*KITSMRRN 57
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 24.3 bits (51), Expect = 7.9
Identities = 8/21 (38%), Positives = 13/21 (61%)
Frame = -2
Query: 56 H*LAHYICVIDHLNKTVHSFI 76
H H +C++ LN +H+FI
Sbjct: 2909 HHSPHCLCMVSSLNARIHNFI 2847
>TC9349 homologue to UP|Q8L3U4 (Q8L3U4) AT5g36790/f5h8_20, partial (34%)
Length = 656
Score = 24.3 bits (51), Expect = 7.9
Identities = 9/20 (45%), Positives = 12/20 (60%)
Frame = +3
Query: 114 LLGKLVWNLLMPTNHLWSQI 133
LLGK +W+ + H W QI
Sbjct: 147 LLGKRIWHFQVTDMHGWGQI 206
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.337 0.146 0.476
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,824,688
Number of Sequences: 28460
Number of extensions: 35483
Number of successful extensions: 325
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 323
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 323
length of query: 136
length of database: 4,897,600
effective HSP length: 81
effective length of query: 55
effective length of database: 2,592,340
effective search space: 142578700
effective search space used: 142578700
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 50 (23.9 bits)
Medicago: description of AC149305.9


