

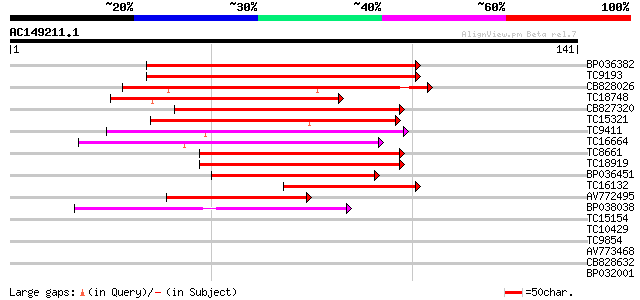
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149211.1 + phase: 0
(141 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP036382 81 8e-17
TC9193 similar to GB|AAN28890.1|23506061|AY143951 At4g02590/T10P... 69 2e-13
CB828026 62 3e-11
TC18748 similar to GB|AAN28890.1|23506061|AY143951 At4g02590/T10... 58 5e-10
CB827320 56 3e-09
TC15321 similar to UP|BAD09664 (BAD09664) BHLH protein family-li... 55 5e-09
TC9411 similar to UP|Q41102 (Q41102) Phaseolin G-box binding pro... 54 1e-08
TC16664 similar to UP|BAD09830 (BAD09830) DNA-binding protein-li... 52 4e-08
TC8661 weakly similar to UP|O81348 (O81348) Symbiotic ammonium t... 49 4e-07
TC18919 similar to GB|AAN73298.1|25141207|BT002301 At4g16430/dl4... 48 7e-07
BP036451 46 3e-06
TC16132 similar to GB|AAN28890.1|23506061|AY143951 At4g02590/T10... 42 4e-05
AV772495 39 3e-04
BP038038 39 4e-04
TC15154 similar to UP|Q9ASX9 (Q9ASX9) F1N18.24/F1N18.24, partial... 33 0.024
TC10429 similar to UP|Q9ZR46 (Q9ZR46) P69C protein, partial (52%) 31 0.071
TC9854 weakly similar to UP|Q9LSN7 (Q9LSN7) Gb|AAC24081.1 (AT3g1... 31 0.092
AV773468 30 0.12
CB828632 30 0.16
BP032001 29 0.35
>BP036382
Length = 511
Score = 80.9 bits (198), Expect = 8e-17
Identities = 38/68 (55%), Positives = 55/68 (80%)
Frame = +3
Query: 35 PRSSSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKTDKASMLDEAIEYLKQLQL 94
PR ++R +A + H+++E+ RR +I E++KALQ L+PN+NKTDKASMLDE I+Y+K LQL
Sbjct: 96 PRVRARRGQATDPHSIAERLRRERIAERMKALQELVPNANKTDKASMLDEIIDYVKFLQL 275
Query: 95 QVQVSFLS 102
QV+V +S
Sbjct: 276 QVKVLSMS 299
>TC9193 similar to GB|AAN28890.1|23506061|AY143951 At4g02590/T10P11_13
{Arabidopsis thaliana;}, partial (59%)
Length = 1474
Score = 69.3 bits (168), Expect = 2e-13
Identities = 31/68 (45%), Positives = 53/68 (77%)
Frame = +1
Query: 35 PRSSSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKTDKASMLDEAIEYLKQLQL 94
PR ++R +A + H+++E+ RR +I E+++A Q L+P+ NKTD+A+MLDE ++Y+K L+L
Sbjct: 427 PRVRARRGQATDPHSIAERLRRERIAERIRA*QELVPSVNKTDRAAMLDEIVDYVKFLRL 606
Query: 95 QVQVSFLS 102
QV+V +S
Sbjct: 607 QVKVLSMS 630
>CB828026
Length = 542
Score = 62.4 bits (150), Expect = 3e-11
Identities = 36/83 (43%), Positives = 56/83 (67%), Gaps = 6/83 (7%)
Frame = +2
Query: 29 SKAAPPPRSS-----SKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNK-TDKASML 82
S+A+ P + ++R +A H+L+E+ RR KI+E++K LQ+L+P +K T KA ML
Sbjct: 77 SQASDPAKEEYIHVRARRGQATNSHSLAERVRREKISERMKFLQDLVPGCSKVTGKAVML 256
Query: 83 DEAIEYLKQLQLQVQVSFLSLKM 105
DE I Y++ LQ QV+ FLS+K+
Sbjct: 257 DEIINYVQSLQRQVE--FLSMKL 319
>TC18748 similar to GB|AAN28890.1|23506061|AY143951 At4g02590/T10P11_13
{Arabidopsis thaliana;}, partial (36%)
Length = 803
Score = 58.2 bits (139), Expect = 5e-10
Identities = 27/60 (45%), Positives = 44/60 (73%), Gaps = 2/60 (3%)
Frame = +1
Query: 26 LPSSKAAPP--PRSSSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKTDKASMLD 83
+PS+ PP PR ++R +A + H+++E+ RR +I E+++ALQ L+P+ NKTD+A MLD
Sbjct: 424 VPSAPHPPPMRPRVRARRGQATDPHSIAERLRRERIAERIRALQELVPSVNKTDRAVMLD 603
>CB827320
Length = 522
Score = 55.8 bits (133), Expect = 3e-09
Identities = 26/57 (45%), Positives = 43/57 (74%)
Frame = +1
Query: 42 SRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKTDKASMLDEAIEYLKQLQLQVQV 98
S AA + +SE+ RR K+NE+L AL++++PN +K DKAS++ +AIEY++ L Q ++
Sbjct: 214 SSAASKNIVSERNRRKKLNERLFALRSVVPNISKMDKASIIKDAIEYIEHLHEQEKI 384
>TC15321 similar to UP|BAD09664 (BAD09664) BHLH protein family-like, partial
(23%)
Length = 1593
Score = 55.1 bits (131), Expect = 5e-09
Identities = 29/63 (46%), Positives = 45/63 (71%), Gaps = 1/63 (1%)
Frame = +2
Query: 36 RSSSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNS-NKTDKASMLDEAIEYLKQLQL 94
+++ K S H+++E+RRRSKINE+ + L+++IP+S K D AS L E IEY++ LQ
Sbjct: 356 KATDKASAIRSKHSVTEQRRRSKINERFQILRDIIPHSEQKRDTASFLLEVIEYVQYLQE 535
Query: 95 QVQ 97
+VQ
Sbjct: 536 KVQ 544
>TC9411 similar to UP|Q41102 (Q41102) Phaseolin G-box binding protein PG2
(Fragment), partial (30%)
Length = 705
Score = 53.5 bits (127), Expect = 1e-08
Identities = 29/82 (35%), Positives = 48/82 (58%), Gaps = 7/82 (8%)
Frame = +2
Query: 25 ELPSSKAAPPPRSSSKRSRAAEF-------HNLSEKRRRSKINEKLKALQNLIPNSNKTD 77
E SS+ P + KR R H +E++RR K+N++ AL+ ++PN +K D
Sbjct: 101 EADSSRLVEPEKRPRKRGRKPANGREEPLNHVEAERQRREKLNQRFYALRAVVPNVSKMD 280
Query: 78 KASMLDEAIEYLKQLQLQVQVS 99
KAS+L +AI Y+ +L+ ++Q S
Sbjct: 281 KASLLGDAISYITELKTKLQSS 346
>TC16664 similar to UP|BAD09830 (BAD09830) DNA-binding protein-like, partial
(22%)
Length = 1067
Score = 52.0 bits (123), Expect = 4e-08
Identities = 30/77 (38%), Positives = 46/77 (58%), Gaps = 1/77 (1%)
Frame = +1
Query: 18 LKNDEGSELPSSKAAPPPRSSSKRS-RAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKT 76
L D+G + + S +RS A H+ +E+RRR++IN L L+ +IP +NK
Sbjct: 199 LDRDKGELVEAPMQLERKGVSPERSIEALRNHSEAERRRRARINAHLDTLRTVIPGANKM 378
Query: 77 DKASMLDEAIEYLKQLQ 93
DKAS+L E I +LK+L+
Sbjct: 379 DKASLLAEVITHLKELK 429
>TC8661 weakly similar to UP|O81348 (O81348) Symbiotic ammonium
transporter, partial (34%)
Length = 1007
Score = 48.5 bits (114), Expect = 4e-07
Identities = 23/51 (45%), Positives = 38/51 (74%)
Frame = +1
Query: 48 HNLSEKRRRSKINEKLKALQNLIPNSNKTDKASMLDEAIEYLKQLQLQVQV 98
H ++E+RRR K+++ L AL LIP K DKAS+L +AI+Y+K L+ ++++
Sbjct: 31 HIIAERRRREKLSQSLIALAALIPGLKKMDKASVLGDAIKYVKVLKERLRL 183
>TC18919 similar to GB|AAN73298.1|25141207|BT002301 At4g16430/dl4240w
{Arabidopsis thaliana;}, partial (29%)
Length = 604
Score = 47.8 bits (112), Expect = 7e-07
Identities = 20/51 (39%), Positives = 38/51 (74%)
Frame = +3
Query: 48 HNLSEKRRRSKINEKLKALQNLIPNSNKTDKASMLDEAIEYLKQLQLQVQV 98
H +E++RR K+N++ AL+ ++PN +K DKAS+L +AI ++ LQ ++++
Sbjct: 414 HVEAERQRREKLNQRFYALRAVVPNISKMDKASLLGDAITHITDLQKKIKL 566
>BP036451
Length = 511
Score = 45.8 bits (107), Expect = 3e-06
Identities = 19/42 (45%), Positives = 33/42 (78%)
Frame = +2
Query: 51 SEKRRRSKINEKLKALQNLIPNSNKTDKASMLDEAIEYLKQL 92
+EK RR ++N K K L++LIP+ K D+AS++ +AI+Y+++L
Sbjct: 332 TEKERREQLNGKYKILRSLIPSPTKIDRASVVGDAIDYIREL 457
>TC16132 similar to GB|AAN28890.1|23506061|AY143951 At4g02590/T10P11_13
{Arabidopsis thaliana;}, partial (32%)
Length = 802
Score = 42.0 bits (97), Expect = 4e-05
Identities = 18/34 (52%), Positives = 29/34 (84%)
Frame = +1
Query: 69 LIPNSNKTDKASMLDEAIEYLKQLQLQVQVSFLS 102
L+P+ NK+D+A+MLDE ++Y+K L+LQV+V +S
Sbjct: 4 LVPSINKSDRAAMLDEIVDYVKFLRLQVKVLSMS 105
>AV772495
Length = 572
Score = 38.9 bits (89), Expect = 3e-04
Identities = 16/36 (44%), Positives = 25/36 (69%)
Frame = -3
Query: 40 KRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNK 75
+R +A + H+L+E+ RR KIN ++K LQ L+P K
Sbjct: 447 RRGQATDSHSLAERARREKINARMKLLQELVPGCYK 340
>BP038038
Length = 464
Score = 38.5 bits (88), Expect = 4e-04
Identities = 21/69 (30%), Positives = 35/69 (50%)
Frame = +1
Query: 17 GLKNDEGSELPSSKAAPPPRSSSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNSNKT 76
G + G+ + PP + + + + H K+R+ K+ E++ ALQ L+ KT
Sbjct: 265 GSGQEPGTTANVGRQKPPYKKTKAENPTSTGH---AKKRKEKLGERIAALQQLVSPXGKT 435
Query: 77 DKASMLDEA 85
D AS+L EA
Sbjct: 436 DTASVLHEA 462
>TC15154 similar to UP|Q9ASX9 (Q9ASX9) F1N18.24/F1N18.24, partial (15%)
Length = 615
Score = 32.7 bits (73), Expect = 0.024
Identities = 16/62 (25%), Positives = 35/62 (55%), Gaps = 1/62 (1%)
Frame = +3
Query: 37 SSSKRSRAAEFHNLSEKRRRSKINEKLKALQNLIPNS-NKTDKASMLDEAIEYLKQLQLQ 95
SSS + + + + ++ ++ L+ ++P ++ D S+LDEA++YLK L+ +
Sbjct: 75 SSSVQKSSGTRGYCNSETNHMEMKNMVRMLRRIVPGGGSQVDAVSVLDEAVKYLKSLKAE 254
Query: 96 VQ 97
V+
Sbjct: 255 VE 260
>TC10429 similar to UP|Q9ZR46 (Q9ZR46) P69C protein, partial (52%)
Length = 1742
Score = 31.2 bits (69), Expect = 0.071
Identities = 16/38 (42%), Positives = 21/38 (55%)
Frame = +3
Query: 3 DYFDNFFYSFLFVFGLKNDEGSELPSSKAAPPPRSSSK 40
DYF +FL F + N +GSELP++ SSSK
Sbjct: 36 DYFLFIALTFLLTFHVHNAQGSELPTTTTESTETSSSK 149
>TC9854 weakly similar to UP|Q9LSN7 (Q9LSN7) Gb|AAC24081.1
(AT3g17100/K14A17_22_), partial (43%)
Length = 1108
Score = 30.8 bits (68), Expect = 0.092
Identities = 18/58 (31%), Positives = 32/58 (55%), Gaps = 1/58 (1%)
Frame = +3
Query: 41 RSRAAEFHNLSEKRRR-SKINEKLKALQNLIPNSNKTDKASMLDEAIEYLKQLQLQVQ 97
RS+ A F L K + + K++ L L+P K +++EAI+Y+ L++QV+
Sbjct: 702 RSKKARFSVLRLKGKTLPAVQRKVRFLGRLVPGCWKEPLPVIVEEAIDYIPALEMQVR 875
>AV773468
Length = 479
Score = 30.4 bits (67), Expect = 0.12
Identities = 13/36 (36%), Positives = 24/36 (66%), Gaps = 1/36 (2%)
Frame = -3
Query: 63 LKALQNLIPNSNKT-DKASMLDEAIEYLKQLQLQVQ 97
++ L+ ++P D S+LDEA++YLK L+ +V+
Sbjct: 465 VRMLRRIVPGGGSPMDAVSVLDEAVKYLKSLKAEVE 358
>CB828632
Length = 365
Score = 30.0 bits (66), Expect = 0.16
Identities = 15/29 (51%), Positives = 18/29 (61%)
Frame = +3
Query: 27 PSSKAAPPPRSSSKRSRAAEFHNLSEKRR 55
PSS AAPPPR S +RA +L+ K R
Sbjct: 27 PSSMAAPPPRPPSP*NRATTSRHLNPKSR 113
>BP032001
Length = 223
Score = 28.9 bits (63), Expect = 0.35
Identities = 12/23 (52%), Positives = 17/23 (73%)
Frame = +2
Query: 31 AAPPPRSSSKRSRAAEFHNLSEK 53
+APPPR+SS R+RA E +S +
Sbjct: 5 SAPPPRASSSRTRATERARMSSR 73
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.132 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,553,614
Number of Sequences: 28460
Number of extensions: 33662
Number of successful extensions: 453
Number of sequences better than 10.0: 82
Number of HSP's better than 10.0 without gapping: 444
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 450
length of query: 141
length of database: 4,897,600
effective HSP length: 81
effective length of query: 60
effective length of database: 2,592,340
effective search space: 155540400
effective search space used: 155540400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 51 (24.3 bits)
Medicago: description of AC149211.1


