

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
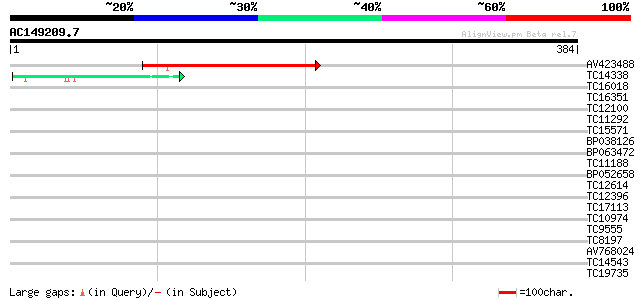
Query= AC149209.7 - phase: 0
(384 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV423488 171 2e-43
TC14338 similar to UP|Q9MAH5 (Q9MAH5) F12M16.16, partial (33%) 44 6e-05
TC16018 UP|Q8MZB8 (Q8MZB8) AT15479p (CG31542-PA), partial (7%) 39 0.002
TC16351 similar to UP|Q84TQ7 (Q84TQ7) GIA/RGA-like gibberellin r... 36 0.009
TC12100 similar to UP|DER7_MOUSE (Q60774) Dermal papilla derived... 33 0.10
TC11292 weakly similar to UP|Q9FLY0 (Q9FLY0) Similarity to ornit... 33 0.10
TC15571 weakly similar to PIR|S19652|S19652 cellodextrinase C - ... 32 0.14
BP038126 32 0.23
BP063472 31 0.40
TC11188 similar to UP|Q84WI7 (Q84WI7) Expressed protein, partial... 31 0.40
BP052658 30 0.88
TC12614 30 0.88
TC12396 similar to UP|Q940Z6 (Q940Z6) At1g12810/F13K23_4, partia... 30 0.88
TC17113 weakly similar to UP|Q9LFU8 (Q9LFU8) Proline-rich protei... 29 1.5
TC10974 29 1.5
TC9555 similar to UP|Q9MUE2 (Q9MUE2) S-adenosyl-L-methionine Mg-... 28 2.0
TC8197 UP|Q8S2Z7 (Q8S2Z7) GTP-binding protein, complete 28 2.0
AV768024 28 2.0
TC14543 similar to UP|Q9ZSY2 (Q9ZSY2) ALTERED response to gravit... 28 2.6
TC19735 similar to UP|Q8NJM5 (Q8NJM5) Probable ATP-dependent RNA... 28 2.6
>AV423488
Length = 487
Score = 171 bits (432), Expect = 2e-43
Identities = 78/122 (63%), Positives = 94/122 (76%), Gaps = 2/122 (1%)
Frame = +3
Query: 91 NGYMPMVNENFQSVG--EYPDFSSQINRGGMTRDNEVAPTPEDTTPKSKRNQQPSWNTEQ 148
+GYM MVNENF SV E+P+FS+Q+ GGMT N+ P EDTTPKSK+ P+W+T Q
Sbjct: 120 SGYMAMVNENFSSVDAPEFPEFSTQMTLGGMTAANQFTPNQEDTTPKSKKTHSPAWSTAQ 299
Query: 149 NLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYCNEHCSFDSPRDIVACRNRFNYMNKLI 208
NLVLIS WI GT SVVG+NQ E++W IAEYCNEHCSFD PRD +ACRNR+NYMNK++
Sbjct: 300 NLVLISRWINRGTSSVVGKNQKGETFWRDIAEYCNEHCSFDPPRDGIACRNRWNYMNKIL 479
Query: 209 NK 210
K
Sbjct: 480 GK 485
>TC14338 similar to UP|Q9MAH5 (Q9MAH5) F12M16.16, partial (33%)
Length = 1751
Score = 43.5 bits (101), Expect = 6e-05
Identities = 41/131 (31%), Positives = 52/131 (39%), Gaps = 15/131 (11%)
Frame = +2
Query: 3 PTNNQSN--TQNSSDYPFSYQ-NQFPNSHPQNPNQFP--NQ----HPQN------PNQFP 47
P +NQ N+ YP S + P P NP +P NQ HPQN PN
Sbjct: 830 PPSNQGGFPPNNAGGYPASPRPGNMPGPPPSNPGGYPPNNQAGGYHPQNQAGGYPPNSQA 1009
Query: 48 NQNPQNMSNFGFASNFNHPSFVPNNFNPYFRSMMGYSSQTPPFNGYMPMVNENFQSVGEY 107
P N G+ N + PNN Y + Y+ +TPP G P N + Y
Sbjct: 1010GGYPPNSQAGGYPPNSQAGVYPPNNQGVYPPNQNNYAGRTPPNVGGAP-PNSAYGPNAGY 1186
Query: 108 PDFSSQINRGG 118
P S+Q N G
Sbjct: 1187P--SNQNNYAG 1213
>TC16018 UP|Q8MZB8 (Q8MZB8) AT15479p (CG31542-PA), partial (7%)
Length = 425
Score = 38.5 bits (88), Expect = 0.002
Identities = 24/88 (27%), Positives = 36/88 (40%)
Frame = +2
Query: 5 NNQSNTQNSSDYPFSYQNQFPNSHPQNPNQFPNQHPQNPNQFPNQNPQNMSNFGFASNFN 64
+N + +SS F + NSH + + N+ + Q PN P S S+FN
Sbjct: 2 SNSNPLASSSTMEFPFNQGRNNSHATHTHHHNNRRDDDEEQHPNYPPPPGSTGSPYSSFN 181
Query: 65 HPSFVPNNFNPYFRSMMGYSSQTPPFNG 92
+P P P++ Q PPF G
Sbjct: 182NPYPPPQQQPPFYGEPHRPPPQQPPFYG 265
>TC16351 similar to UP|Q84TQ7 (Q84TQ7) GIA/RGA-like gibberellin response
modulator, partial (11%)
Length = 756
Score = 36.2 bits (82), Expect = 0.009
Identities = 21/63 (33%), Positives = 33/63 (52%), Gaps = 2/63 (3%)
Frame = +1
Query: 25 PNSHPQNPNQFP--NQHPQNPNQFPNQNPQNMSNFGFASNFNHPSFVPNNFNPYFRSMMG 82
P P +P+ FP + HP +P+ P +P S+ + NF P+F + NP+F S +
Sbjct: 7 PYLFPSDPSPFPLWSPHPSHPSSQPTNSP*LFSSILKSHNFT*PNF---DSNPFFTSSIS 177
Query: 83 YSS 85
SS
Sbjct: 178SSS 186
>TC12100 similar to UP|DER7_MOUSE (Q60774) Dermal papilla derived protein
7 homolog (19.5), partial (7%)
Length = 573
Score = 32.7 bits (73), Expect = 0.10
Identities = 14/42 (33%), Positives = 25/42 (59%)
Frame = +1
Query: 4 TNNQSNTQNSSDYPFSYQNQFPNSHPQNPNQFPNQHPQNPNQ 45
T+ ++ ++S P S+++Q P+ H NP F + Q+PNQ
Sbjct: 49 TSRFFSSASASTEPNSHRSQIPHRHHHNPQLFLSHRSQHPNQ 174
>TC11292 weakly similar to UP|Q9FLY0 (Q9FLY0) Similarity to ornithine
cyclodeaminase, partial (47%)
Length = 551
Score = 32.7 bits (73), Expect = 0.10
Identities = 20/52 (38%), Positives = 24/52 (45%), Gaps = 1/52 (1%)
Frame = +2
Query: 28 HPQNPNQ-FPNQHPQNPNQFPNQNPQNMSNFGFASNFNHPSFVPNNFNPYFR 78
HPQN +Q PNQ PQ+ N PN N + F H V F P+ R
Sbjct: 131 HPQNSHQSLPNQSPQSFNLSPNPNSPKLQCLSFFLPPPHAFLVFFPFLPFHR 286
>TC15571 weakly similar to PIR|S19652|S19652 cellodextrinase C - Pseudomonas
fluorescens {Pseudomonas fluorescens;} , partial (6%)
Length = 784
Score = 32.3 bits (72), Expect = 0.14
Identities = 14/31 (45%), Positives = 19/31 (61%)
Frame = +3
Query: 21 QNQFPNSHPQNPNQFPNQHPQNPNQFPNQNP 51
+NQ P HP NQFP H +P++ NQ+P
Sbjct: 687 KNQSPPHHPDQMNQFPPHHHHHPDRL-NQSP 776
>BP038126
Length = 548
Score = 31.6 bits (70), Expect = 0.23
Identities = 14/28 (50%), Positives = 16/28 (57%)
Frame = +2
Query: 25 PNSHPQNPNQFPNQHPQNPNQFPNQNPQ 52
P+ P NP QF HP P +FPN N Q
Sbjct: 185 PSRSPSNPPQF-QLHPSTPEKFPNPNIQ 265
>BP063472
Length = 459
Score = 30.8 bits (68), Expect = 0.40
Identities = 16/34 (47%), Positives = 19/34 (55%), Gaps = 4/34 (11%)
Frame = -3
Query: 24 FPNSHPQNPNQFPNQHPQNPNQ----FPNQNPQN 53
FP H QNP PN HPQ+ Q FP ++ QN
Sbjct: 217 FPQKHKQNPISQPN-HPQSKQQLLLLFPQKHKQN 119
>TC11188 similar to UP|Q84WI7 (Q84WI7) Expressed protein, partial (18%)
Length = 523
Score = 30.8 bits (68), Expect = 0.40
Identities = 11/21 (52%), Positives = 17/21 (80%)
Frame = -1
Query: 28 HPQNPNQFPNQHPQNPNQFPN 48
+PQ P FP Q+PQ+P++FP+
Sbjct: 127 YPQLPLLFPGQNPQSPHRFPS 65
>BP052658
Length = 522
Score = 29.6 bits (65), Expect = 0.88
Identities = 19/65 (29%), Positives = 28/65 (42%), Gaps = 9/65 (13%)
Frame = +2
Query: 35 FP-NQHPQNPNQFPNQNPQNMSNFGFASNFNHP----SFVPN----NFNPYFRSMMGYSS 85
FP NQHPQ+ P+ + S AS+ HP +F+P + P +R +
Sbjct: 290 FPSNQHPQSTTNIPDSVALDASKISSASSIRHPAPAHTFIPTMPLPSEPPNYRXATSWMX 469
Query: 86 QTPPF 90
P F
Sbjct: 470 TPPTF 484
>TC12614
Length = 577
Score = 29.6 bits (65), Expect = 0.88
Identities = 11/23 (47%), Positives = 14/23 (60%)
Frame = +3
Query: 30 QNPNQFPNQHPQNPNQFPNQNPQ 52
Q P FP Q+P +PN P +PQ
Sbjct: 123 QTPTSFPTQNPNSPNPKPMASPQ 191
Score = 29.3 bits (64), Expect = 1.1
Identities = 18/55 (32%), Positives = 28/55 (50%), Gaps = 4/55 (7%)
Frame = +2
Query: 25 PNSHPQNPNQFPNQHPQNP----NQFPNQNPQNMSNFGFASNFNHPSFVPNNFNP 75
PNS NP+ FPN P P N P++NP+ ++ + N + + P+N P
Sbjct: 107 PNSTLPNPHLFPNPKP*FPQSKTNGVPSRNPR*TTSSSPSPNPSTNTPSPSNLTP 271
Score = 26.6 bits (57), Expect = 7.5
Identities = 12/32 (37%), Positives = 14/32 (43%)
Frame = +3
Query: 10 TQNSSDYPFSYQNQFPNSHPQNPNQFPNQHPQ 41
T N PF FP +P +PN P PQ
Sbjct: 96 TFNHRIRPFQTPTSFPTQNPNSPNPKPMASPQ 191
>TC12396 similar to UP|Q940Z6 (Q940Z6) At1g12810/F13K23_4, partial (39%)
Length = 408
Score = 29.6 bits (65), Expect = 0.88
Identities = 22/84 (26%), Positives = 34/84 (40%), Gaps = 14/84 (16%)
Frame = +3
Query: 48 NQNPQNMSNFGFASNFNHPSFVPNNFN---------PYFRSMMGYSSQTPP--FNGY--- 93
+Q PQ MSN+ A +P + P + P + GY S PP + GY
Sbjct: 33 SQIPQTMSNYQRAPQEPYPDYPPPGYGSPYPPPQGYPVTQPPPGYPSAPPPPGYEGYPPP 212
Query: 94 MPMVNENFQSVGEYPDFSSQINRG 117
P V + + +Y + N+G
Sbjct: 213 PPPVYAGYAARPQYEGYQGYFNQG 284
>TC17113 weakly similar to UP|Q9LFU8 (Q9LFU8) Proline-rich protein, partial
(25%)
Length = 1077
Score = 28.9 bits (63), Expect = 1.5
Identities = 19/57 (33%), Positives = 25/57 (43%), Gaps = 3/57 (5%)
Frame = +1
Query: 22 NQFPNSHPQ---NPNQFPNQHPQNPNQFPNQNPQNMSNFGFASNFNHPSFVPNNFNP 75
N F S P +P Q P+ P PN F +P + F + P F+PN F P
Sbjct: 508 NPFQPSPPPLLPDPFQPPSPPPLFPNPFQPPSPPPLIPNPFQPPPSPPPFIPNPFQP 678
>TC10974
Length = 652
Score = 28.9 bits (63), Expect = 1.5
Identities = 20/70 (28%), Positives = 28/70 (39%), Gaps = 15/70 (21%)
Frame = +3
Query: 22 NQFPNSHPQNP----NQFPNQHPQN-------PNQFPNQNPQNMSNFGFASNFNHPSF-- 68
NQ N PQ P NQ + P PN PN + +N+ ++FN P
Sbjct: 75 NQMGNPMPQGPFVGMNQMHSGPPSGGPPLGGFPNHLPNMQGPSNTNYPQGASFNRPQGGQ 254
Query: 69 --VPNNFNPY 76
+ +NPY
Sbjct: 255 MPMMQGYNPY 284
>TC9555 similar to UP|Q9MUE2 (Q9MUE2) S-adenosyl-L-methionine
Mg-protoporphyrin IX methyltranserase , partial (34%)
Length = 586
Score = 28.5 bits (62), Expect = 2.0
Identities = 12/26 (46%), Positives = 15/26 (57%)
Frame = +2
Query: 25 PNSHPQNPNQFPNQHPQNPNQFPNQN 50
PNSH Q + FP+ PQ P P Q+
Sbjct: 221 PNSHSQPHSPFPHSQPQPPPTSPAQS 298
>TC8197 UP|Q8S2Z7 (Q8S2Z7) GTP-binding protein, complete
Length = 996
Score = 28.5 bits (62), Expect = 2.0
Identities = 34/160 (21%), Positives = 62/160 (38%), Gaps = 22/160 (13%)
Frame = +3
Query: 85 SQTPPFNGYMPMVNENFQSVGEYPDFSSQINRGGMTR-------------DNEVAPT--- 128
S + P N +PM N Q+V +YP+F I G T + + PT
Sbjct: 60 SFSSPINTALPMALPNQQTV-DYPNFKLVIVGDGGTGKTTFVKRHLTGEFEKKYEPTIGV 236
Query: 129 ---PEDTTPKSKRNQQPSWNT---EQNLVLISGWIKYGTCSVVGRNQTSESYWGKIAEYC 182
P D + + W+T E+ L G+ +G C+++ + T+ + + +
Sbjct: 237 EVHPLDFFTNCGKIRFYCWDTAGQEKFGGLRDGYYIHGQCAIIMFDVTARLTYKNVPTWH 416
Query: 183 NEHCSFDSPRDIVACRNRFNYMNKLINKWVGAYDSAKRMQ 222
+ C IV C N+ + N+ + + K +Q
Sbjct: 417 RDLCRVCENIPIVLCGNKVDVKNRQVKAKQVTFHRKKNLQ 536
>AV768024
Length = 439
Score = 28.5 bits (62), Expect = 2.0
Identities = 32/124 (25%), Positives = 52/124 (41%), Gaps = 5/124 (4%)
Frame = +2
Query: 12 NSSDYPFSYQNQFPNSHPQNPN-----QFPNQHPQNPNQFPNQNPQNMSNFGFASNFNHP 66
N+ D F +++ P + P NP+ Q P+ HP +FP P
Sbjct: 41 NNKDLSFDAKSKPPTNFP-NPSMLSQMQSPSLHPFKSLKFP-------------IPLITP 178
Query: 67 SFVPNNFNPYFRSMMGYSSQTPPFNGYMPMVNENFQSVGEYPDFSSQINRGGMTRDNEVA 126
+PN+ +P+ S+ +S TPP P+++E Q P S+I + +
Sbjct: 179 PSLPNSPSPF--SLRVRTSTTPP----PPLISE--QQTPNSPSTQSRIAAAPPSTNLGFR 334
Query: 127 PTPE 130
PTPE
Sbjct: 335 PTPE 346
>TC14543 similar to UP|Q9ZSY2 (Q9ZSY2) ALTERED response to gravity (ARG1
protein), partial (50%)
Length = 1078
Score = 28.1 bits (61), Expect = 2.6
Identities = 23/61 (37%), Positives = 29/61 (46%), Gaps = 1/61 (1%)
Frame = +1
Query: 259 QPRYGSQVGGNVDSGSSGSKRSYEDSVGSS-ARPMDRDAAKKKGKKKSKGETLEKVEKEW 317
+P S G N+ +GSS SK S EDS G S D K GKKK L+ +K
Sbjct: 535 KPTNISGSGSNLSNGSS-SKISGEDSKGESPGEEGGSDGKDKSGKKKWFNLNLKSSDKRL 711
Query: 318 V 318
+
Sbjct: 712 I 714
>TC19735 similar to UP|Q8NJM5 (Q8NJM5) Probable ATP-dependent RNA helicase,
partial (5%)
Length = 565
Score = 28.1 bits (61), Expect = 2.6
Identities = 16/39 (41%), Positives = 25/39 (64%), Gaps = 2/39 (5%)
Frame = +3
Query: 282 EDSVGSSARPMDRDAAKKKGKKKSKGETLEKVEK--EWV 318
E+ +G+++ D +++KKK KKK K E E+ EK WV
Sbjct: 255 EEQIGNNS-DRDGESSKKKKKKKHKVEVEERKEKNNSWV 368
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.311 0.129 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,083,577
Number of Sequences: 28460
Number of extensions: 115643
Number of successful extensions: 841
Number of sequences better than 10.0: 81
Number of HSP's better than 10.0 without gapping: 755
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 822
length of query: 384
length of database: 4,897,600
effective HSP length: 92
effective length of query: 292
effective length of database: 2,279,280
effective search space: 665549760
effective search space used: 665549760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC149209.7


