

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149130.7 - phase: 0 /pseudo
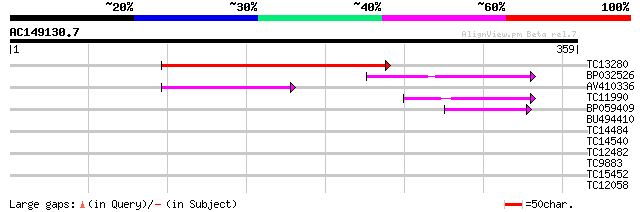
(359 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC13280 weakly similar to GB|AAP68292.1|31711872|BT008853 At5g11... 140 3e-34
BP032526 70 5e-13
AV410336 66 8e-12
TC11990 49 1e-06
BP059409 47 4e-06
BU494410 28 1.8
TC14484 similar to UP|SGPL_ARATH (Q9C509) Sphingosine-1-phosphat... 28 2.4
TC14540 similar to UP|O49855 (O49855) Acid phosphatase , partial... 28 2.4
TC12482 similar to UP|Q9TD47 (Q9TD47) NADH dehydrogenase subunit... 27 5.3
TC9883 homologue to UP|AOX2_SOYBN (Q41266) Alternative oxidase 2... 27 5.3
TC15452 similar to UP|SR40_YEAST (P32583) Suppressor protein SRP... 26 9.0
TC12058 homologue to GB|AAO64878.1|29028998|BT005943 At5g37010 {... 26 9.0
>TC13280 weakly similar to GB|AAP68292.1|31711872|BT008853 At5g11730
{Arabidopsis thaliana;}, partial (39%)
Length = 702
Score = 140 bits (353), Expect = 3e-34
Identities = 67/146 (45%), Positives = 100/146 (67%), Gaps = 1/146 (0%)
Frame = +3
Query: 97 PKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHASKA-KPVHVSRYFVNRDIRSDQLV 155
PK+AF+FL ++P LW+ FF+GHEG FS+YVH+ + S F R I S +
Sbjct: 264 PKVAFLFLVRSNVPLAPLWEVFFRGHEGYFSIYVHSHPSYNGSDKSPLFRGRRIPSKIVE 443
Query: 156 WGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRDP 215
WG+++++EAERRLLANAL D +NQ FVL+S+SC+PL+NF+ I+ YLM + +++V + +P
Sbjct: 444 WGRVNMMEAERRLLANALLDFSNQRFVLISESCIPLFNFSTIYSYLMNSTQNYVMAVDEP 623
Query: 216 GPVGNGRYSEHMLPEVEIKDFRTGAQ 241
G GRY M PE+ ++ +R G+Q
Sbjct: 624 SAAGRGRYKIQMSPEITLRQWRKGSQ 701
>BP032526
Length = 516
Score = 70.1 bits (170), Expect = 5e-13
Identities = 38/108 (35%), Positives = 61/108 (56%), Gaps = 1/108 (0%)
Frame = -1
Query: 227 MLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPT 286
MLPEV FR G+Q+F L R+HAV V+ D + KF+ C+ + C +EHY PT
Sbjct: 498 MLPEVTFDAFRVGSQFFVLTRRHAVVVLRDRRLWRKFRL----PCLTEEPCYPEEHYFPT 331
Query: 287 FFTIVDPNGIAKWSVTYVDRSE-QKRHPKSYRTQDITYELLKNIKLHN 333
++ DPNG +++T V+ + HP Y +++ EL++ ++ N
Sbjct: 330 LLSMEDPNGCTGFTLTRVNWTGCWDGHPHLYTPPEVSPELVRRLRESN 187
>AV410336
Length = 358
Score = 66.2 bits (160), Expect = 8e-12
Identities = 36/87 (41%), Positives = 51/87 (58%), Gaps = 2/87 (2%)
Frame = +3
Query: 97 PKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHASKAKP-VHVSRYFVNRDIRSDQLV 155
PKIAF+FLT +L F LW+ FF GH +++Y+HA + V F R I S +
Sbjct: 96 PKIAFLFLTNSNLTFAPLWEKFFTGHHHLYNIYIHADPSSAVVSPGGVFHRRFITSKKTY 275
Query: 156 WGKMSIVEAERRLLANA-LQDPNNQHF 181
+++ A RRLLA+A L DP N++F
Sbjct: 276 RSSPTLISATRRLLASAVLDDPLNEYF 356
>TC11990
Length = 544
Score = 49.3 bits (116), Expect = 1e-06
Identities = 26/84 (30%), Positives = 42/84 (49%)
Frame = +3
Query: 250 AVKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQ 309
A++V++D Y+ F+ C C DEHYLPT I + ++T+VD ++
Sbjct: 3 ALQVVSDRRYFPVFEKYCNPQCYS------DEHYLPTLVNIKFWKRNSNRTLTWVDWTKG 164
Query: 310 KRHPKSYRTQDITYELLKNIKLHN 333
HP Y D+T E L+ ++ N
Sbjct: 165 GPHPSKYIRTDVTVEFLERLRYGN 236
>BP059409
Length = 357
Score = 47.4 bits (111), Expect = 4e-06
Identities = 22/55 (40%), Positives = 32/55 (58%)
Frame = -1
Query: 276 NCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELLKNIK 330
+C DEHYLPTF I+ + S+T+VD S HP S+ D+T E L+ ++
Sbjct: 354 SCYADEHYLPTFVNIMFGERNSNRSLTWVDWSRGGPHPASFTRSDVTVEFLERLR 190
>BU494410
Length = 310
Score = 28.5 bits (62), Expect = 1.8
Identities = 12/38 (31%), Positives = 21/38 (54%)
Frame = +2
Query: 249 HAVKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPT 286
H +++ A+H S Q QC+Q + NC ++ H + T
Sbjct: 95 HMMRIPANHSCMSSMQKQCQQILLHL*NCRIEFHMVST 208
>TC14484 similar to UP|SGPL_ARATH (Q9C509) Sphingosine-1-phosphate lyase
(SP-lyase) (SPL) (Sphingosine-1-phosphate aldolase) ,
partial (20%)
Length = 725
Score = 28.1 bits (61), Expect = 2.4
Identities = 17/51 (33%), Positives = 24/51 (46%)
Frame = +2
Query: 171 NALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRDPGPVGNG 221
NALQ PN+ H CV L + D+L +S ++PGP+ G
Sbjct: 143 NALQRPNSIHI------CVTLQLVPAVDDFLKDLKESVETVKKNPGPISGG 277
>TC14540 similar to UP|O49855 (O49855) Acid phosphatase , partial (88%)
Length = 1144
Score = 28.1 bits (61), Expect = 2.4
Identities = 15/33 (45%), Positives = 17/33 (51%)
Frame = -1
Query: 201 LMYTDKSFVDSFRDPGPVGNGRYSEHMLPEVEI 233
LM DKS + SFRD G G S H L V +
Sbjct: 544 LMPRDKSLLYSFRDSGSTGAAARSTHRLNAVSL 446
>TC12482 similar to UP|Q9TD47 (Q9TD47) NADH dehydrogenase subunit II,
partial (5%)
Length = 720
Score = 26.9 bits (58), Expect = 5.3
Identities = 16/55 (29%), Positives = 26/55 (47%)
Frame = -3
Query: 149 IRSDQLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMY 203
I+ D V KM +++ +LL N + D + F+ L N +F YL+Y
Sbjct: 403 IKCDNFV--KMVVIKTNPKLLLNGVVDLQHFCFIKTIIKVCLLRNITRVFSYLLY 245
>TC9883 homologue to UP|AOX2_SOYBN (Q41266) Alternative oxidase 2,
mitochondrial precursor , partial (34%)
Length = 665
Score = 26.9 bits (58), Expect = 5.3
Identities = 14/54 (25%), Positives = 28/54 (50%)
Frame = +3
Query: 227 MLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDGKNCILD 280
+L + E +D++ G +L+ HAV ++ L + +A C + G +C+ D
Sbjct: 345 LLGDFEAEDYQRGRHRVALELLHAVGDLSSRLVHRSDEASCAEE-FSG*SCLQD 503
>TC15452 similar to UP|SR40_YEAST (P32583) Suppressor protein SRP40, partial
(4%)
Length = 589
Score = 26.2 bits (56), Expect = 9.0
Identities = 11/22 (50%), Positives = 13/22 (59%)
Frame = -2
Query: 330 KLHNFHLFLTERSTKMDLFLEW 351
+L NF LFL R+T FL W
Sbjct: 285 RLSNFTLFLAVRTTGFPFFLSW 220
>TC12058 homologue to GB|AAO64878.1|29028998|BT005943 At5g37010 {Arabidopsis
thaliana;}, partial (5%)
Length = 426
Score = 26.2 bits (56), Expect = 9.0
Identities = 16/67 (23%), Positives = 29/67 (42%), Gaps = 8/67 (11%)
Frame = +2
Query: 289 TIVDPNGIAKWSVTYVDRSEQKR--------HPKSYRTQDITYELLKNIKLHNFHLFLTE 340
T++ P+G+ + RS R +P++ +TY L +HNFH T+
Sbjct: 74 TVIVPSGVDNHKPQTLTRSRSSRRSSRDLDINPEALLNPPLTYTSLLLEDIHNFHQKNTQ 253
Query: 341 RSTKMDL 347
+ + L
Sbjct: 254 QQPSVSL 274
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.325 0.139 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,102,608
Number of Sequences: 28460
Number of extensions: 105241
Number of successful extensions: 691
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 687
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 688
length of query: 359
length of database: 4,897,600
effective HSP length: 91
effective length of query: 268
effective length of database: 2,307,740
effective search space: 618474320
effective search space used: 618474320
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC149130.7


