

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149129.2 + phase: 0
(261 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
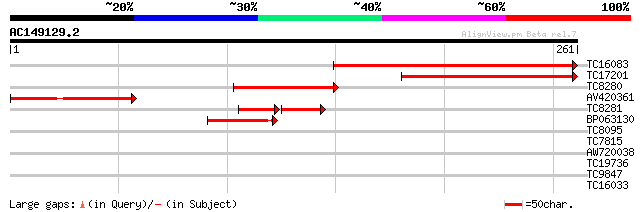
Score E
Sequences producing significant alignments: (bits) Value
TC16083 similar to UP|Q940L9 (Q940L9) At2g44770/F16B22.26, parti... 162 4e-41
TC17201 similar to UP|Q940L9 (Q940L9) At2g44770/F16B22.26, parti... 115 1e-26
TC8280 81 2e-16
AV420361 73 6e-14
TC8281 similar to UP|Q940L9 (Q940L9) At2g44770/F16B22.26, partia... 45 3e-12
BP063130 59 6e-10
TC8095 32 0.11
TC7815 similar to UP|Q9AVB5 (Q9AVB5) Plasma membrane intrinsic p... 28 1.6
AW720038 28 1.6
TC19736 homologue to UP|RL34_PEA (P40590) 60S ribosomal protein ... 27 2.7
TC9847 similar to UP|AAR14272 (AAR14272) Predicted protein, part... 27 3.5
TC16033 UP|Q7X9C1 (Q7X9C1) NIN-like protein 1, partial (88%) 26 7.9
>TC16083 similar to UP|Q940L9 (Q940L9) At2g44770/F16B22.26, partial (42%)
Length = 788
Score = 162 bits (411), Expect = 4e-41
Identities = 84/112 (75%), Positives = 89/112 (79%)
Frame = +1
Query: 150 SRKEIVQCGSTRLRLLVLTLHSCLFKC*ILKQLSHGHWWEQLS*NSLQKMSRPLIFSIA* 209
SRKE QCG+T L LLVLT HSCLF+C ILKQ SHGH EQLS N LQK ++ LIFSIA
Sbjct: 4 SRKEFDQCGNTHLLLLVLT*HSCLFRCWILKQSSHGHL*EQLSFNFLQKTNQLLIFSIAL 183
Query: 210 HSS*WTINGFPCTRHTWILIQ**NLHAVSWRKNFSLKTLHN*KMYPHTNFSH 261
SS*W INGF C HTWILIQ**NLHAVSWR +F LKTLH KMYPHTNFSH
Sbjct: 184 RSS*WIINGFLCAHHTWILIQ**NLHAVSWRMSFCLKTLHGLKMYPHTNFSH 339
>TC17201 similar to UP|Q940L9 (Q940L9) At2g44770/F16B22.26, partial (32%)
Length = 576
Score = 115 bits (287), Expect = 1e-26
Identities = 59/81 (72%), Positives = 66/81 (80%)
Frame = +2
Query: 181 QLSHGHWWEQLS*NSLQKMSRPLIFSIA*HSS*WTINGFPCTRHTWILIQ**NLHAVSWR 240
Q SH HW EQLS*N LQKM++ LI SIA* SS*W INGF C HTWILIQ**NLH VSWR
Sbjct: 14 Q*SHTHW*EQLS*NFLQKMNQLLICSIA*LSS*WIINGFLCVPHTWILIQ**NLHDVSWR 193
Query: 241 KNFSLKTLHN*KMYPHTNFSH 261
K++SLKT H+ K+YPHTN+ H
Sbjct: 194 KSYSLKT*HSLKIYPHTNYFH 256
>TC8280
Length = 502
Score = 80.9 bits (198), Expect = 2e-16
Identities = 36/48 (75%), Positives = 41/48 (85%)
Frame = +1
Query: 104 LNGLISEQWKDMGWQGKDPSTDFRGGGYISLENLLFFARNFPIFCGSR 151
L+GLIS+QWK+M WQ KDPSTDFRGGG+ISLEN LFF+RNFP SR
Sbjct: 1 LHGLISDQWKEMDWQAKDPSTDFRGGGFISLENFLFFSRNFPELKRSR 144
>AV420361
Length = 250
Score = 72.8 bits (177), Expect = 6e-14
Identities = 37/58 (63%), Positives = 44/58 (75%)
Frame = +1
Query: 1 MDDRGGSFVAVRRISQGLDRGNAYHSSSAEFVTGSTAWIGRGLSCVCAQRRESDARLS 58
MDDRGGSFVAVRR+ G + +S+SAE V GS AW+G+GLSCVC QRR+SDA S
Sbjct: 82 MDDRGGSFVAVRRVPHGETCHS--NSNSAEAVAGSAAWLGKGLSCVCVQRRDSDASSS 249
>TC8281 similar to UP|Q940L9 (Q940L9) At2g44770/F16B22.26, partial (14%)
Length = 509
Score = 45.4 bits (106), Expect(2) = 3e-12
Identities = 18/19 (94%), Positives = 19/19 (99%)
Frame = +2
Query: 106 GLISEQWKDMGWQGKDPST 124
GLISEQWK+MGWQGKDPST
Sbjct: 2 GLISEQWKEMGWQGKDPST 58
Score = 41.6 bits (96), Expect(2) = 3e-12
Identities = 17/20 (85%), Positives = 19/20 (95%)
Frame = +1
Query: 126 FRGGGYISLENLLFFARNFP 145
FRGGG+ISLEN LFF+RNFP
Sbjct: 61 FRGGGFISLENFLFFSRNFP 120
>BP063130
Length = 504
Score = 59.3 bits (142), Expect = 6e-10
Identities = 26/32 (81%), Positives = 30/32 (93%)
Frame = -2
Query: 92 RALWNAAFPEEELNGLISEQWKDMGWQGKDPS 123
RALWNAAFPEE+L+GLIS+QWK+MGWQ DPS
Sbjct: 104 RALWNAAFPEEKLHGLISDQWKEMGWQA-DPS 12
>TC8095
Length = 1147
Score = 32.0 bits (71), Expect = 0.11
Identities = 30/108 (27%), Positives = 46/108 (41%)
Frame = +1
Query: 147 FCGSRKEIVQCGSTRLRLLVLTLHSCLFKC*ILKQLSHGHWWEQLS*NSLQKMSRPLIFS 206
+C ++ E Q G+ L+ L C +C I KQ H + +S + L++M
Sbjct: 601 YCINKMEPEQSGNIHLQ*LESIFLLCWRRCWIFKQGIHLLCQDSVSCSCLRRMKWHSTSF 780
Query: 207 IA*HSS*WTINGFPCTRHTWILIQ**NLHAVSWRKNFSLKTLHN*KMY 254
I *W ++G H W L *+ S + LKT *K+Y
Sbjct: 781 IVLPIK*WMLSG*QSVLHIWNLTMF*SPQERS*SVSLLLKTSPG*KIY 924
>TC7815 similar to UP|Q9AVB5 (Q9AVB5) Plasma membrane intrinsic protein
2-1, complete
Length = 1450
Score = 28.1 bits (61), Expect = 1.6
Identities = 15/41 (36%), Positives = 21/41 (50%)
Frame = +3
Query: 10 AVRRISQGLDRGNAYHSSSAEFVTGSTAWIGRGLSCVCAQR 50
AV R D+G + HSS+ F WIG+GL + Q+
Sbjct: 390 AVHRTQGVFDKGTSIHSSTV-FRCNQWCWIGQGLPKIILQQ 509
>AW720038
Length = 561
Score = 28.1 bits (61), Expect = 1.6
Identities = 20/50 (40%), Positives = 28/50 (56%), Gaps = 1/50 (2%)
Frame = +2
Query: 29 AEFVTGSTAWIGRGLSCVC-AQRRESDARLSFDLTPYQEECLQRLQSRID 77
A V GS + IGR LS A +++ L LTP QEE L+ L+ R++
Sbjct: 410 ANIVLGSGSLIGRLLSVSSTALYLQNNRMLP*PLTPLQEERLRNLRQRLE 559
>TC19736 homologue to UP|RL34_PEA (P40590) 60S ribosomal protein L34,
partial (98%)
Length = 612
Score = 27.3 bits (59), Expect = 2.7
Identities = 16/62 (25%), Positives = 26/62 (41%)
Frame = +3
Query: 64 YQEECLQRLQSRIDVPYDSSIPEHQASLRALWNAAFPEEELNGLISEQWKDMGWQGKDPS 123
++E+ +QRL R Y + +H W A P+ + G QW + K+ S
Sbjct: 9 HEEKMVQRLTYRKRHSYATKSNQHXGRQNPRWEARLPDHQEEG----QWTQVPCHWKEDS 176
Query: 124 TD 125
D
Sbjct: 177 GD 182
>TC9847 similar to UP|AAR14272 (AAR14272) Predicted protein, partial (73%)
Length = 952
Score = 26.9 bits (58), Expect = 3.5
Identities = 13/34 (38%), Positives = 19/34 (55%)
Frame = -3
Query: 26 SSSAEFVTGSTAWIGRGLSCVCAQRRESDARLSF 59
S+ A + ST+WI C C++ RES + SF
Sbjct: 656 STRARLLRSSTSWI-----CKCSRHRESSSSCSF 570
>TC16033 UP|Q7X9C1 (Q7X9C1) NIN-like protein 1, partial (88%)
Length = 2554
Score = 25.8 bits (55), Expect = 7.9
Identities = 17/63 (26%), Positives = 29/63 (45%), Gaps = 1/63 (1%)
Frame = +3
Query: 60 DLTPYQEECLQRLQSRIDVPYDSSIPEHQASLRALWNA-AFPEEELNGLISEQWKDMGWQ 118
DLT + +Q + D Y++++PE Q LR+ + P +++ W Q
Sbjct: 591 DLTSLKHSSIQNAKQARDKSYEAALPEIQEVLRSACHMHKLP-------LAQTWVSCFQQ 749
Query: 119 GKD 121
GKD
Sbjct: 750 GKD 758
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.337 0.145 0.501
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,697,934
Number of Sequences: 28460
Number of extensions: 90904
Number of successful extensions: 580
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 579
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 579
length of query: 261
length of database: 4,897,600
effective HSP length: 88
effective length of query: 173
effective length of database: 2,393,120
effective search space: 414009760
effective search space used: 414009760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 54 (25.4 bits)
Medicago: description of AC149129.2


