

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149127.4 + phase: 0
(323 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
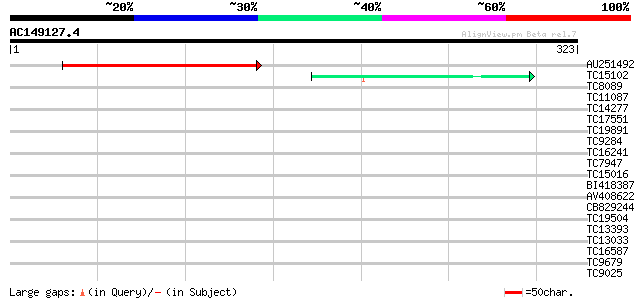
Score E
Sequences producing significant alignments: (bits) Value
AU251492 135 9e-33
TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, ... 40 7e-04
TC8089 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinog... 39 0.001
TC11087 weakly similar to UP|Q8GS92 (Q8GS92) OJ1513_F02.5 protei... 35 0.017
TC14277 similar to UP|Q9SHY3 (Q9SHY3) F1E22.9 (AT1G65720/F1E22_1... 35 0.017
TC17551 weakly similar to UP|AAR24662 (AAR24662) At5g26960, part... 35 0.022
TC19891 homologue to PIR|S14182|S14182 DNA-directed RNA polymera... 34 0.038
TC9284 weakly similar to UP|Q9LPY0 (Q9LPY0) T23J18.22, partial (... 33 0.050
TC16241 weakly similar to UP|Q91TH4 (Q91TH4) T122, partial (7%) 33 0.050
TC7947 similar to UP|Q9ST69 (Q9ST69) Ribosomal protein S5, parti... 33 0.050
TC15016 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pro... 33 0.065
BI418387 33 0.085
AV408622 33 0.085
CB829244 32 0.15
TC19504 similar to UP|Q9SV89 (Q9SV89) Isoleucine-tRNA ligase-lik... 32 0.15
TC13393 similar to UP|Q9FMU6 (Q9FMU6) Mitochondrial phosphate tr... 32 0.19
TC13033 weakly similar to PIR|JC4847|JC4847 DNA-directed RNA pol... 32 0.19
TC16587 weakly similar to UP|Q9JGS5 (Q9JGS5) PORF1, partial (3%) 32 0.19
TC9679 weakly similar to UP|Q8H1B5 (Q8H1B5) Hin1-like protein, p... 32 0.19
TC9025 similar to PIR|F86282|F86282 protein F10B6.30 [imported] ... 31 0.25
>AU251492
Length = 393
Score = 135 bits (340), Expect = 9e-33
Identities = 64/113 (56%), Positives = 86/113 (75%)
Frame = +2
Query: 31 LTDLKNLRTQLYSAAEYFELSYTNDDQKQILVETLKDYAVKALINTVDHLGSVAYKVSDL 90
L +LKNLR QLYSAAEY E SY + +QKQ++++ LKDYAV+AL+N VDHLG+VAYK++DL
Sbjct: 2 LQELKNLRPQLYSAAEYCEKSYLHSEQKQMVLDNLKDYAVRALVNAVDHLGTVAYKLTDL 181
Query: 91 LDEKVTEVFGEDLRLSCIEQRIKTCQGFMDHEGHTQQSLVISTPKHHKRYILP 143
LD+ +V DLR+S + Q++ TC+ + D EG QQ L+ P+HHK YILP
Sbjct: 182 LDQHTFDVSSMDLRVSTVNQKLLTCKIYTDKEGLRQQQLLAFIPRHHKHYILP 340
>TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, partial
(68%)
Length = 1097
Score = 39.7 bits (91), Expect = 7e-04
Identities = 30/134 (22%), Positives = 49/134 (36%), Gaps = 7/134 (5%)
Frame = +1
Query: 173 RNAVRATIRETPTSTSSKGNSPSPSLQP-------QRVGAFSFTSPNMAKKDLEKRTVSP 225
+NA T P S +P+PS P + + + PN L SP
Sbjct: 361 KNATSLTAPSCPPPASPAATAPAPSKTPPNPPTNSSKPSSCKSSLPNTTPSSLSSAPPSP 540
Query: 226 HRFPLSRTGSMSSRSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVN 285
P+S S ++ T S P S++ +++SP N P +P + SS
Sbjct: 541 TTSPISTPSSPENQEKPASTPPSAPPRSTSSSSSSPINT----PRKPSSATKAPASSPPG 708
Query: 286 NIRDIDQHPSKSKR 299
+ + P + R
Sbjct: 709 SELSLHHSPPSASR 750
Score = 31.2 bits (69), Expect = 0.25
Identities = 26/95 (27%), Positives = 41/95 (42%)
Frame = +1
Query: 183 TPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTT 242
TP+S SS PSP+ P ++P+ + + + P P S + S SS T
Sbjct: 505 TPSSLSSA--PPSPTTSP-------ISTPSSPENQEKPASTPPSAPPRSTSSSSSSPINT 657
Query: 243 PKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSAS 277
P+ S T ++ S + PS RKS++
Sbjct: 658 PRKPSSATKAPASSPPGSELSLHHSPPSASRKSST 762
Score = 26.6 bits (57), Expect = 6.1
Identities = 24/111 (21%), Positives = 43/111 (38%), Gaps = 3/111 (2%)
Frame = +1
Query: 156 SKYIGCHLDDEDDWHHFRNAVRATIRETPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAK 215
++Y + W T T S + SPS +P + T+
Sbjct: 37 NRYSSVRSSHQQQWQQHLQTTPKTAAATFL*NPSLEATASPSSEPCTTATTTSTTKAATN 216
Query: 216 KDL-EKRTVSPHRFPLSRTGSMSSRST--TPKTGRSTTPNSSNRATTSPSN 263
E R+ +P P S + + ++ TPK+ S+TP S ++T+P +
Sbjct: 217 SSPPESRSTTP---PSSEPTCLLAPASPPTPKSSHSSTPPPSRSSSTTPKS 360
>TC8089 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinogalactan
protein 1 precursor, partial (43%)
Length = 890
Score = 38.9 bits (89), Expect = 0.001
Identities = 28/93 (30%), Positives = 43/93 (46%), Gaps = 2/93 (2%)
Frame = +2
Query: 183 TPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTT 242
TP+S S SPSP+ P+ S + + H+ P SRT S S+ S+T
Sbjct: 257 TPSSPPSTTTSPSPTSPPRSTNGPQSPSAPSTTPPWTTSSQNTHQSPPSRTSSPSTSSST 436
Query: 243 PKTGRSTTPNSSNRATTSPSNAR--VRYPSEPR 273
++ P SS R+ T+P + + P+ PR
Sbjct: 437 -----TSAPRSSTRSPTAPPSPPQCTKPPAPPR 520
>TC11087 weakly similar to UP|Q8GS92 (Q8GS92) OJ1513_F02.5 protein
(OJ1118_G09.13 protein), partial (6%)
Length = 689
Score = 35.0 bits (79), Expect = 0.017
Identities = 40/139 (28%), Positives = 59/139 (41%), Gaps = 22/139 (15%)
Frame = +1
Query: 180 IRETPTSTSSKG-------NSPSPSL----QPQRVGAFSFTSPNMAKKDLEKRTVSPHRF 228
I + P+ST SK ++PSPS +P ++P K T SP
Sbjct: 1 INQNPSSTCSKTTTPMATWSTPSPSTPTPSKPSSPQIPPTSTPGSPKPPATATTNSP--- 171
Query: 229 PLSRTGSMSSRSTTPKTGR----------STTPNSSNRATTSPSNARVRYPSEPRKSASM 278
S + S SS + TPK R STT SS++++T P + + PS P + +
Sbjct: 172 --SSSVSTSSGAPTPKPSRTTPSPSSNSASTTAASSSKSSTHPPSLTLCLPSSPTPNTRL 345
Query: 279 -RLSSDVNNIRDIDQHPSK 296
L S V +I PS+
Sbjct: 346 *ALGSRVTSISFSRTGPSR 402
>TC14277 similar to UP|Q9SHY3 (Q9SHY3) F1E22.9 (AT1G65720/F1E22_13), partial
(33%)
Length = 941
Score = 35.0 bits (79), Expect = 0.017
Identities = 37/113 (32%), Positives = 46/113 (39%), Gaps = 5/113 (4%)
Frame = +3
Query: 157 KYIGCHLDDEDDWHHFRNAVRATIRETPTSTSSKGNSPSPSLQPQRVGAFSFTSPN-MAK 215
K++ L D D W + + +P S+SS SPSP P A S + P A
Sbjct: 33 KFLYISLSDSDPW-----LLSSAASRSPPSSSS---SPSPPPPPHAPAAASSSPPTPSAT 188
Query: 216 KDLEKRTVSPHRFP----LSRTGSMSSRSTTPKTGRSTTPNSSNRATTSPSNA 264
SP P S T +S STTPK TT N SN T S+A
Sbjct: 189 LPPTPSPPSPRSDPSPLSTSTTNPTTSSSTTPK----TTTNKSNPNTARLSHA 335
>TC17551 weakly similar to UP|AAR24662 (AAR24662) At5g26960, partial (22%)
Length = 505
Score = 34.7 bits (78), Expect = 0.022
Identities = 30/95 (31%), Positives = 41/95 (42%), Gaps = 1/95 (1%)
Frame = +3
Query: 183 TPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTT 242
T STSS + P PSL A TS + P S + +S S
Sbjct: 147 TSFSTSSPASHPLPSLPSPSSAAVGPTSSTPPTSPTSAAAATSSATPPSPSPPPTSASPP 326
Query: 243 PKTGR-STTPNSSNRATTSPSNARVRYPSEPRKSA 276
P + + TP SS+ ATT PS +R PS P +++
Sbjct: 327 PPSSTPNGTPPSSSPATT-PSPSRPSTPSSPTRAS 428
>TC19891 homologue to PIR|S14182|S14182 DNA-directed RNA polymerase largest
chain (isoform B2) - soybean (fragment)
{Glycine max;} , partial (34%)
Length = 498
Score = 33.9 bits (76), Expect = 0.038
Identities = 29/76 (38%), Positives = 36/76 (47%), Gaps = 1/76 (1%)
Frame = +3
Query: 187 TSSKGNSP-SPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPKT 245
TSS G SP SP P G +S TSP + SP P S T S SS +P T
Sbjct: 276 TSSPGYSPSSPGYSPSSPG-YSPTSPGYSPTSPGYSPTSPGYSPTSPTYSPSSPGYSP-T 449
Query: 246 GRSTTPNSSNRATTSP 261
+ +P S + + TSP
Sbjct: 450 SPAYSPTSPSYSPTSP 497
>TC9284 weakly similar to UP|Q9LPY0 (Q9LPY0) T23J18.22, partial (33%)
Length = 898
Score = 33.5 bits (75), Expect = 0.050
Identities = 27/100 (27%), Positives = 42/100 (42%)
Frame = +1
Query: 183 TPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTT 242
TPT+T+ + + P + S T P+ A K PH S + S STT
Sbjct: 145 TPTTTNESASFSPSNPDPNPASSASTTKPSAATSPPSK----PHAPTPSSSSSTPLSSTT 312
Query: 243 PKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSS 282
S + +S+ ++ PS+ P R S+S L+S
Sbjct: 313 NPNSASPSFATSSPSSPPPSSPTTPTPPHSRISSSATLNS 432
>TC16241 weakly similar to UP|Q91TH4 (Q91TH4) T122, partial (7%)
Length = 799
Score = 33.5 bits (75), Expect = 0.050
Identities = 28/104 (26%), Positives = 44/104 (41%), Gaps = 3/104 (2%)
Frame = +1
Query: 177 RATIRETPTSTSSKGNSPSPSLQPQRVGAFSFTSP---NMAKKDLEKRTVSPHRFPLSRT 233
RA + S+S NSP PS P + TSP A + P + P + +
Sbjct: 226 RAVTPISSPSSSPTSNSPYPSTNPPSLAPSISTSPPALTPASTPVPATASPPSQSPSAES 405
Query: 234 GSMSSRSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSAS 277
S + T + STTP R+T +PS+ P++ ++S
Sbjct: 406 PGASLPAATTPSPSSTTP----RSTQTPSSNNETTPAQRNGASS 525
Score = 28.5 bits (62), Expect = 1.6
Identities = 25/96 (26%), Positives = 42/96 (43%), Gaps = 2/96 (2%)
Frame = +1
Query: 184 PTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEK--RTVSPHRFPLSRTGSMSSRST 241
P +T + N+PSPS QP + ++ ++ + + R V+P P S S S +
Sbjct: 109 PPNTQTPANTPSPSSQPPYLSTPPKSNSPTSQPPVAQTPRAVTPISSPSSSPTSNSPYPS 288
Query: 242 TPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSAS 277
T + + ++S A T S S P +S S
Sbjct: 289 TNPPSLAPSISTSPPALTPASTPVPATASPPSQSPS 396
>TC7947 similar to UP|Q9ST69 (Q9ST69) Ribosomal protein S5, partial (81%)
Length = 1294
Score = 33.5 bits (75), Expect = 0.050
Identities = 27/106 (25%), Positives = 50/106 (46%), Gaps = 10/106 (9%)
Frame = +2
Query: 183 TPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTT 242
+P S S+ + PSPS S +SP+ + P+ P + +++ S+T
Sbjct: 134 SPLSPSAPPH-PSPSSPQPPNSPSSLSSPDPSSPPPPPPPSPPNANPPT---TLTPPSST 301
Query: 243 PKTGRSTTPNSSNRA----------TTSPSNARVRYPSEPRKSASM 278
P+T + T+P++ ++ TT+PS R + P RKS ++
Sbjct: 302 PQTQKKTSPSTHQKSPKASSLLLPSTTAPSKPRTKSPPRMRKSTAL 439
>TC15016 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (4%)
Length = 948
Score = 33.1 bits (74), Expect = 0.065
Identities = 23/96 (23%), Positives = 41/96 (41%), Gaps = 3/96 (3%)
Frame = +1
Query: 183 TPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTT 242
TPT TSS + P+ + +SP + S P ++ S S+STT
Sbjct: 73 TPTKTSSFSSQPTTTTTAAAAQPPPASSPPSSSSSSSPPPPSSSTHPTRKSASSESKSTT 252
Query: 243 PKTGRSTTPNSSNRATTS---PSNARVRYPSEPRKS 275
+ + +P+S++R+ + + YP+ P S
Sbjct: 253 SGSESTRSPSSTSRSPSEFKFATGTSSPYPTTPSPS 360
>BI418387
Length = 479
Score = 32.7 bits (73), Expect = 0.085
Identities = 29/100 (29%), Positives = 42/100 (42%), Gaps = 3/100 (3%)
Frame = +2
Query: 186 STSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRT-VSP--HRFPLSRTGSMSSRSTT 242
STS+ + S +P + TSP+ L RT +SP P + T S S T
Sbjct: 158 STSTNSSESHYSTKPCNTIISTETSPSTCIPSLPFRTLISPTSSLAPPNTTSSSPSAPTK 337
Query: 243 PKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSS 282
P T S+ P + PS++R P+ SA +S
Sbjct: 338 PSTTTSSEPPFPGPTSPQPSSSRSENPTGAESSAPTSTTS 457
>AV408622
Length = 415
Score = 32.7 bits (73), Expect = 0.085
Identities = 41/142 (28%), Positives = 57/142 (39%), Gaps = 2/142 (1%)
Frame = +1
Query: 140 YILPVGET--LHGTNSTKSKYIGCHLDDEDDWHHFRNAVRATIRETPTSTSSKGNSPSPS 197
Y+LP +T LH K D HH R R+ P+ST+S P PS
Sbjct: 40 YLLPASKTEALHSVEDNTLKR-----DPFLHHHHLRLRFRSPPPWPPSSTNSHSLQPRPS 204
Query: 198 LQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPKTGRSTTPNSSNRA 257
PQ + P+ A +SP + G++SS + +P+ P N A
Sbjct: 205 SAPQSI----LHPPSSA--------LSPPHPIGNPFGTLSSVAPSPRKHPRQNP-PRNPA 345
Query: 258 TTSPSNARVRYPSEPRKSASMR 279
T+ PS A P+ SAS R
Sbjct: 346 TSPPSTASSSAPAS-AASASPR 408
>CB829244
Length = 478
Score = 32.0 bits (71), Expect = 0.15
Identities = 36/115 (31%), Positives = 48/115 (41%), Gaps = 17/115 (14%)
Frame = +2
Query: 170 HHFRNAVRATIRETPTSTSSKGNSPSPSLQPQRVGAFSFTSPN---MAKKDLEKRTVSPH 226
HH R V T+ T + P PS +FS T+P+ + + KR VS
Sbjct: 38 HHGRRRVPVTVTVTAAAAP-----P*PSSF-----SFSTTTPSSISLGNTNSSKRCVSFS 187
Query: 227 RFPLSRTG-----SMSSRSTTPKTGRSTTPNSSN---------RATTSPSNARVR 267
P S + S SS +TTP STT +S++ ATT S AR R
Sbjct: 188 STPSSSSSASFPRSSSSTNTTPSRSSSTTSSSTSTRQQHHPTLHATTPQSGARCR 352
>TC19504 similar to UP|Q9SV89 (Q9SV89) Isoleucine-tRNA ligase-like protein
, partial (11%)
Length = 491
Score = 32.0 bits (71), Expect = 0.15
Identities = 33/100 (33%), Positives = 46/100 (46%)
Frame = +3
Query: 184 PTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTP 243
P S SK +SPSP + SP + L T SP + S+T SS +T+P
Sbjct: 144 PESMPSKPSSPSPRTCRSTSSTTAHPSPPAS---LTTATSSPAQ---SKT---SSPATSP 296
Query: 244 KTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSSD 283
++ S+ AT SPS R SE R S+R+ S+
Sbjct: 297 *PATTSRAASAGTATASPSRTRSTRSSESR---SVRMCSN 407
>TC13393 similar to UP|Q9FMU6 (Q9FMU6) Mitochondrial phosphate translocator
(AT5g14040/MUA22_4), partial (8%)
Length = 541
Score = 31.6 bits (70), Expect = 0.19
Identities = 23/92 (25%), Positives = 42/92 (45%), Gaps = 6/92 (6%)
Frame = +1
Query: 184 PTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSM------S 237
P S SS +P P+ +SP+ + + + FP++ TGS+ S
Sbjct: 58 PPSASSTFPNPPPTRT---------SSPSSSPHSVPPPSTPSKSFPITPTGSLAAPAASS 210
Query: 238 SRSTTPKTGRSTTPNSSNRATTSPSNARVRYP 269
SR++ P S +PN + +++ S+A + P
Sbjct: 211 SRTSPPSPAPSLSPNPRSSSSSPTSSASPKTP 306
>TC13033 weakly similar to PIR|JC4847|JC4847 DNA-directed RNA polymerase
chain III - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (73%)
Length = 594
Score = 31.6 bits (70), Expect = 0.19
Identities = 20/49 (40%), Positives = 27/49 (54%)
Frame = +2
Query: 149 HGTNSTKSKYIGCHLDDEDDWHHFRNAVRATIRETPTSTSSKGNSPSPS 197
HG++S +SK L DED H F NA+R T+ + P T + P PS
Sbjct: 113 HGSHSDESKST-FSLTDED--HTFANAIRYTLNQDPRVTFCGYSIPHPS 250
>TC16587 weakly similar to UP|Q9JGS5 (Q9JGS5) PORF1, partial (3%)
Length = 706
Score = 31.6 bits (70), Expect = 0.19
Identities = 26/92 (28%), Positives = 42/92 (45%), Gaps = 6/92 (6%)
Frame = +1
Query: 192 NSPSPSLQPQRV-GAFSFTSPNMAKKDLEKRT--VSPHRFPLSRTGSMSSRSTTPKTGR- 247
+S +P ++P + G FT + R S RF + + S+ +++P T R
Sbjct: 136 HSQNPQIKPNQT*GFIFFTK*KLLSNSATNRRSHCSEPRFHSASSACRSNPASSPATPRN 315
Query: 248 --STTPNSSNRATTSPSNARVRYPSEPRKSAS 277
ST+P+SSN A S + P P S+S
Sbjct: 316 SHSTSPHSSNLALPSEPLTALTTPKTPSPSSS 411
>TC9679 weakly similar to UP|Q8H1B5 (Q8H1B5) Hin1-like protein, partial
(54%)
Length = 710
Score = 31.6 bits (70), Expect = 0.19
Identities = 36/135 (26%), Positives = 59/135 (43%), Gaps = 17/135 (12%)
Frame = +3
Query: 171 HFRNAVRATIRETPTSTSSKGNSPSPSLQPQR-------VGAFSFTSPNMAKKDLEKRTV 223
H N+ R +I P T ++ + PSL+P++ A + ++ A +
Sbjct: 24 HN*NSARFSIHP-PCQT*TELITAPPSLRPKKPTTAQAEAAADASSAAAAASSASSSNSS 200
Query: 224 SP----HRFPLSRTGSMS------SRSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPR 273
SP P S +GS S S S TP++ ST+P ++ T SPS + PS +
Sbjct: 201 SPPSSSSASPSSSSGSSSAPTS*SSTSPTPRSRSSTSPETTRSTTISPSTS----PSATQ 368
Query: 274 KSASMRLSSDVNNIR 288
SAS ++ +R
Sbjct: 369 TSASGSTTTTSKRVR 413
>TC9025 similar to PIR|F86282|F86282 protein F10B6.30 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(55%)
Length = 991
Score = 31.2 bits (69), Expect = 0.25
Identities = 26/79 (32%), Positives = 33/79 (40%)
Frame = -1
Query: 233 TGSMSSRSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDIDQ 292
T S SSR T + T P+ R TSP PR S S R SS IR
Sbjct: 595 TSSPSSRDRTRRPSPDTPPHPGTRPCTSPRR------*APRSSTSARCSSGTRTIR---- 446
Query: 293 HPSKSKRLLKSLLSRRKSK 311
+ +R L+S+ S S+
Sbjct: 445 -CRRRRRRLQSVASASASR 392
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.313 0.128 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,041,225
Number of Sequences: 28460
Number of extensions: 68488
Number of successful extensions: 616
Number of sequences better than 10.0: 117
Number of HSP's better than 10.0 without gapping: 577
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 598
length of query: 323
length of database: 4,897,600
effective HSP length: 90
effective length of query: 233
effective length of database: 2,336,200
effective search space: 544334600
effective search space used: 544334600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 55 (25.8 bits)
Medicago: description of AC149127.4


