

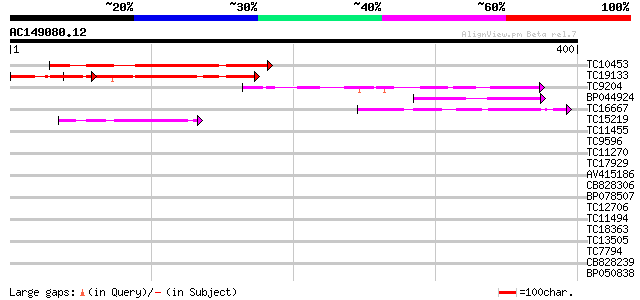
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149080.12 - phase: 0
(400 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC10453 similar to UP|SSGP_VOLCA (P21997) Sulfated surface glyco... 199 9e-52
TC19133 similar to UP|Q7Z697 (Q7Z697) KAISO-like zinc finger pro... 113 6e-26
TC9204 similar to PIR|T00986|T00986 yeast pheromone receptor-lik... 75 2e-14
BP044924 65 3e-11
TC16667 similar to UP|O64594 (O64594) F17O7.4 (AT1G70420/F17O7_4... 50 7e-07
TC15219 similar to PIR|T00986|T00986 yeast pheromone receptor-li... 40 7e-04
TC11455 similar to UP|Q9ZUS8 (Q9ZUS8) At2g37380 protein, partial... 36 0.013
TC9596 similar to UP|AAR24722 (AAR24722) At5g49400, partial (66%) 36 0.013
TC11270 similar to UP|Q9LM88 (Q9LM88) F2D10.15, partial (9%) 35 0.022
TC17929 34 0.038
AV415186 34 0.049
CB828306 33 0.064
BP078507 33 0.11
TC12706 UP|Q7X9C0 (Q7X9C0) NIN-like protein 2, complete 32 0.19
TC11494 similar to UP|Q7X9B2 (Q7X9B2) 9/13 hydroperoxide lyase, ... 32 0.24
TC18363 UP|Q56662 (Q56662) TcpI protein (Fragment), partial (9%) 31 0.32
TC13505 similar to UP|Q9T0K5 (Q9T0K5) Extensin-like protein, par... 31 0.42
TC7794 homologue to UP|Q9M600 (Q9M600) Chlorophyll a/b binding p... 30 0.54
CB828239 30 0.54
BP050838 30 0.54
>TC10453 similar to UP|SSGP_VOLCA (P21997) Sulfated surface glycoprotein 185
precursor (SSG 185), partial (5%)
Length = 461
Score = 199 bits (505), Expect = 9e-52
Identities = 109/157 (69%), Positives = 120/157 (76%)
Frame = +2
Query: 29 DSNCSTPYVSAPSSPGRGGGPPPISSGYFYSAPASPLHFSITASSSYHHQTTTSSVPMSY 88
DS CSTPYVSAPSSP +G++YSAPASP+HF+ SSS
Sbjct: 74 DSTCSTPYVSAPSSP----------AGFYYSAPASPMHFAAITSSS-------------- 181
Query: 89 EFEFSARFGSTGSAASGSMTSADELFLNGQIRPMKLSSHLERPQVLAPLLDLEEEEEDEE 148
EFEFSARFGS A+SG+MTSADELFLNGQIRPMKLS+HLERPQ+LAPLLDLE EEEDEE
Sbjct: 182 EFEFSARFGSAALASSGAMTSADELFLNGQIRPMKLSTHLERPQLLAPLLDLEGEEEDEE 361
Query: 149 EGEVVVVRGRDLRLRDKSVRRRTRSMSPLRNNSHLEW 185
VVVRGRDLRLRDKSVRRRTRSMSPLRN++ LEW
Sbjct: 362 ----VVVRGRDLRLRDKSVRRRTRSMSPLRNDTPLEW 460
>TC19133 similar to UP|Q7Z697 (Q7Z697) KAISO-like zinc finger protein 1,
partial (3%)
Length = 524
Score = 113 bits (282), Expect = 6e-26
Identities = 79/141 (56%), Positives = 87/141 (61%), Gaps = 3/141 (2%)
Frame = -3
Query: 39 APSSPGRGGGPPPISSGYFYSAPASPLHFSITA--SSSYHHQTTTSSVPMSYEFEFSARF 96
AP++ P P SS + P PLHF+ITA SSS Q SS S EFE
Sbjct: 387 APAAAAEVNHPSPASST---APPPGPLHFAITAAASSSSSFQNPFSSTSSS-EFEVLCPV 220
Query: 97 G-STGSAASGSMTSADELFLNGQIRPMKLSSHLERPQVLAPLLDLEEEEEDEEEGEVVVV 155
TG A GSM+SADELF NGQIRPMKL +HLERPQ LLDLEEEE +EE V
Sbjct: 219 RLCTGFGAPGSMSSADELFWNGQIRPMKLPTHLERPQ----LLDLEEEEGEEE----TEV 64
Query: 156 RGRDLRLRDKSVRRRTRSMSP 176
RGRD RLR KS RRRTRS+SP
Sbjct: 63 RGRDWRLRAKSGRRRTRSLSP 1
Score = 63.5 bits (153), Expect = 6e-11
Identities = 38/62 (61%), Positives = 42/62 (67%), Gaps = 1/62 (1%)
Frame = -2
Query: 1 MELQNSTKNPNLEPSNSNNGGETFFEDIDSNCSTPYVSAPSSPG-RGGGPPPISSGYFYS 59
MELQ+ T N + SN + D DS CSTPYVSAPSSPG RGGG PPI SG+FYS
Sbjct: 490 MELQSGTNNGETQTSNLE------YID-DSTCSTPYVSAPSSPGRRGGGEPPI-SGFFYS 335
Query: 60 AP 61
AP
Sbjct: 334 AP 329
>TC9204 similar to PIR|T00986|T00986 yeast pheromone receptor-like protein
AR781 [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (25%)
Length = 644
Score = 75.1 bits (183), Expect = 2e-14
Identities = 70/217 (32%), Positives = 100/217 (45%), Gaps = 4/217 (1%)
Frame = +2
Query: 165 KSVRRRTRSMSPLRNNSHLEWTENEDDNNNKNNVACEIEDEMNNKNNENGNDCSEMENVK 224
KS RR TRS+SP R SH W E E + + E +D +
Sbjct: 29 KSGRRVTRSLSPYRV-SHYTWEE---------------EHQQRQRTKEEKSDLNS----- 145
Query: 225 VDEMGLERIEITPLDSASSSR--SSSAGRSSKRWVFLKDFL--RSKSEGRSNNKFWSTIS 280
P +ASSS S+S G S ++W L+DFL RS SEGR
Sbjct: 146 -----------APATAASSSSLSSTSKGSSGRKWR-LRDFLLFRSASEGRG--------- 262
Query: 281 FSPTKDKKSSSNNQNLQAQISKEKTCETQRNGSSSSSSSSSKGSWGKKMTGKPMNGVGKR 340
SS + + + +K E + +SSS+S+S +GS ++ +R
Sbjct: 263 ---------SSTDPLRKFHLFYKKGEEVK---ASSSTSTSFRGSETPRL---------RR 379
Query: 341 RVPPSPHELHYKANRAQAEELRRKTFLPYRQGLLGCL 377
+ P S HELHY RA+ E+L+++TFLPY+QG+LG L
Sbjct: 380 KEPVSAHELHYARKRAETEDLKKRTFLPYKQGILGRL 490
>BP044924
Length = 561
Score = 64.7 bits (156), Expect = 3e-11
Identities = 39/93 (41%), Positives = 49/93 (51%)
Frame = -2
Query: 286 DKKSSSNNQNLQAQISKEKTCETQRNGSSSSSSSSSKGSWGKKMTGKPMNGVGKRRVPPS 345
DK++ + L + + E RN S S+ SS V KRR P S
Sbjct: 542 DKEALRKYEVLPRKSAATAAEEDVRNCSFRSTESSGS--------------VSKRRGPVS 405
Query: 346 PHELHYKANRAQAEELRRKTFLPYRQGLLGCLG 378
HELHY NRA AEE++R+T LPY+QGL GCLG
Sbjct: 404 AHELHYTLNRAAAEEMKRRTQLPYKQGLFGCLG 306
>TC16667 similar to UP|O64594 (O64594) F17O7.4 (AT1G70420/F17O7_4), partial
(23%)
Length = 953
Score = 50.1 bits (118), Expect = 7e-07
Identities = 44/151 (29%), Positives = 64/151 (42%)
Frame = +2
Query: 246 SSSAGRSSKRWVFLKDFLRSKSEGRSNNKFWSTISFSPTKDKKSSSNNQNLQAQISKEKT 305
SSSA + + KS ++K W +D K SN+ A + T
Sbjct: 422 SSSAAAPEETYCEWSPKTAVKSNSTGSSKVWKF------RDVKLRSNSDGKDAFVFLNST 583
Query: 306 CETQRNGSSSSSSSSSKGSWGKKMTGKPMNGVGKRRVPPSPHELHYKANRAQAEELRRKT 365
++ +S G KK+ GK S HE HY NRA+ E +RK+
Sbjct: 584 ---------AAEKASPTGVVVKKVKAVK----GKTTTSTSAHEKHYVLNRAKKESDKRKS 724
Query: 366 FLPYRQGLLGCLGFSSKGYGAMNGFARALNP 396
+LPYRQ L G GF + + NG ++L+P
Sbjct: 725 YLPYRQELFG--GF----FASTNGMHKSLHP 799
>TC15219 similar to PIR|T00986|T00986 yeast pheromone receptor-like protein
AR781 [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (11%)
Length = 571
Score = 40.0 bits (92), Expect = 7e-04
Identities = 34/102 (33%), Positives = 47/102 (45%)
Frame = +2
Query: 35 PYVSAPSSPGRGGGPPPISSGYFYSAPASPLHFSITASSSYHHQTTTSSVPMSYEFEFSA 94
PY+SAPSSP R G ++ SAP SP F S T S++ +
Sbjct: 221 PYLSAPSSPKRFG------EDFYLSAPTSPSSFY-----SQFDYFTGSNLDGDCDGGDDG 367
Query: 95 RFGSTGSAASGSMTSADELFLNGQIRPMKLSSHLERPQVLAP 136
S G + SA+ELF G+I+P+ SS R +L+P
Sbjct: 368 FAFSVGGGSETPSRSAEELFHGGKIKPLHESS---RTPLLSP 484
>TC11455 similar to UP|Q9ZUS8 (Q9ZUS8) At2g37380 protein, partial (10%)
Length = 575
Score = 35.8 bits (81), Expect = 0.013
Identities = 23/64 (35%), Positives = 32/64 (49%)
Frame = +1
Query: 71 ASSSYHHQTTTSSVPMSYEFEFSARFGSTGSAASGSMTSADELFLNGQIRPMKLSSHLER 130
AS SY +T S S +FEF+ S S ++ ADELF GQ+ P+ LS +
Sbjct: 100 ASKSYTLPSTPSHSSSSSDFEFTISISPRKS--STALCPADELFYKGQLLPLHLSPRISM 273
Query: 131 PQVL 134
+ L
Sbjct: 274 VRTL 285
>TC9596 similar to UP|AAR24722 (AAR24722) At5g49400, partial (66%)
Length = 1196
Score = 35.8 bits (81), Expect = 0.013
Identities = 38/156 (24%), Positives = 70/156 (44%), Gaps = 3/156 (1%)
Frame = +2
Query: 177 LRNNSHLEWTENEDDNNNKNNVACEIEDEMNNKNNENGNDCSEMEN-VKVDEMGLERIEI 235
LR N + + +D ++K A + K+ + + S+ E+ V E G +
Sbjct: 536 LRVNVSVSYDLENNDVDDKEEKAKSSSKKAKRKHRSDSDSGSDSEDSVFETESGSSSVTG 715
Query: 236 TPLDSASSSRSSSAGRSSKRWVFLKDFLRSKSEGRSNNKFWSTISFSPTKDKKSSSNNQN 295
+ S SSS SSS+ +R K + K +GR + K+ S+ S + D +S+S++ +
Sbjct: 716 SDYSSGSSSDSSSSDSEEERRRRRKKKTQKKKKGRRHKKYSSS---SESSDSESASDSDS 886
Query: 296 LQAQISKEKTCETQRN--GSSSSSSSSSKGSWGKKM 329
+ S+ K ++R+ SS G W K+
Sbjct: 887 -DDKSSRRKRRHSRRH*IRLKDVRSSYQFGEWVDKL 991
>TC11270 similar to UP|Q9LM88 (Q9LM88) F2D10.15, partial (9%)
Length = 735
Score = 35.0 bits (79), Expect = 0.022
Identities = 31/113 (27%), Positives = 51/113 (44%), Gaps = 3/113 (2%)
Frame = +3
Query: 140 LEEEEEDEEEGEVVVVRGRDLRLRDKSVRRRTRSMSPLRNNSHLEWTENEDDNNNKNNVA 199
+EEEEEDEE+ E ++ P R S +E + E+D +N
Sbjct: 336 VEEEEEDEEDEE-------------------EQTPHPTRGRSRVEHAQ-EEDEEEENGEE 455
Query: 200 CEIEDEM---NNKNNENGNDCSEMENVKVDEMGLERIEITPLDSASSSRSSSA 249
E E+E+ + + NE+ ++ E E V V E+ +++ L S S + S A
Sbjct: 456 EEEEEEVVVEDEQGNEDEDEDEEEEGVVVVEVKGRKVDSKGLHSGSGTPSKVA 614
Score = 26.6 bits (57), Expect = 7.9
Identities = 22/70 (31%), Positives = 30/70 (42%), Gaps = 9/70 (12%)
Frame = -3
Query: 265 SKSEGRSNNKFWSTISFSPTKDKKSSSNNQNLQAQISKEKTCETQR---------NGSSS 315
S S S++ F + S T SSS++ + S C T+ + SSS
Sbjct: 529 SSSSSSSSSSFPCS---SSTTTSSSSSSSSPFSSSSSSSCACSTRDLPRVGCGVCSSSSS 359
Query: 316 SSSSSSKGSW 325
SSSSSS W
Sbjct: 358 SSSSSSSTPW 329
>TC17929
Length = 791
Score = 34.3 bits (77), Expect = 0.038
Identities = 26/86 (30%), Positives = 44/86 (50%), Gaps = 14/86 (16%)
Frame = +3
Query: 264 RSKSEGRSNNKFWSTIS-----------FSPTKDKKSSSNNQNLQAQISKEKTCETQRNG 312
RS++E S N+ W+ IS +S ++ +S ++++L SK ++ R+G
Sbjct: 381 RSRNESESPNRSWAKISKWSDSSPVPADYSASRSLSNSQSSKSLARPTSKYRS----RSG 548
Query: 313 SSSSSSSSSK---GSWGKKMTGKPMN 335
SS + SSSSK S + GK +N
Sbjct: 549 SSRAHSSSSKLSSSSKSRTYRGKSLN 626
>AV415186
Length = 276
Score = 33.9 bits (76), Expect = 0.049
Identities = 22/67 (32%), Positives = 33/67 (48%)
Frame = +1
Query: 2 ELQNSTKNPNLEPSNSNNGGETFFEDIDSNCSTPYVSAPSSPGRGGGPPPISSGYFYSAP 61
E++ ++P+ P ++ F+ S+PY+SAPSSP R G Y+ SAP
Sbjct: 34 EMEVVVQSPSSPPMDN-------FDFSSGTMSSPYLSAPSSPKRFG-------EYYLSAP 171
Query: 62 ASPLHFS 68
SP S
Sbjct: 172TSPSRIS 192
>CB828306
Length = 550
Score = 33.5 bits (75), Expect = 0.064
Identities = 35/106 (33%), Positives = 48/106 (45%), Gaps = 5/106 (4%)
Frame = -3
Query: 225 VDEMGLERIEITPLDSASSSRSSSAGRSSKRWVFLKDFLRSKSEGRSNNKFWSTISFSPT 284
++ MGL R + L SSS SS + + LK L SKS S + +S SPT
Sbjct: 482 LEGMGLSRFCLNTLYLYSSSSLSS*PCAYTQHQILKAVLGSKSIAESCDIGTVVLSSSPT 303
Query: 285 KD----KKSSSNNQNLQAQISKEKTCETQRNGSSSSSS-SSSKGSW 325
K K S + +++ S KT T +S + S SSS SW
Sbjct: 302 KSFDFAKSMSFSTESVLCSSSPTKTLSTLNFFTSGAKSFSSSTESW 165
>BP078507
Length = 414
Score = 32.7 bits (73), Expect = 0.11
Identities = 26/84 (30%), Positives = 40/84 (46%), Gaps = 2/84 (2%)
Frame = -3
Query: 27 DIDSNCSTPYVSAPSS--PGRGGGPPPISSGYFYSAPASPLHFSITASSSYHHQTTTSSV 84
++ +CS + AP+ P PPP + GY ++ SP T SSS+ + TTS+
Sbjct: 247 NLSLSCSLVPIEAPTIHFPSAPTDPPPTNLGYPFTLSTSPFVTKSTTSSSF-TKPTTSTF 71
Query: 85 PMSYEFEFSARFGSTGSAASGSMT 108
+S AR SA G++T
Sbjct: 70 FLSISSSILAR-----SALHGTVT 14
>TC12706 UP|Q7X9C0 (Q7X9C0) NIN-like protein 2, complete
Length = 3814
Score = 32.0 bits (71), Expect = 0.19
Identities = 31/140 (22%), Positives = 55/140 (39%), Gaps = 8/140 (5%)
Frame = +3
Query: 199 ACEIEDEMNNKNNENGNDCSEMENVKVDEMGLERIEITPLDSASSSRSSSAGRSSKR--W 256
A ++E ++ N + C + +++ +D+ + SS ++S +KR W
Sbjct: 2535 AIKLEGKLKKTNASLVDCCEDSKSMAMDDRSSHSSSLWTKAQGSSEQASLGSDLAKRDKW 2714
Query: 257 VFLKDFLRSKS-----EGRSNNKFWST-ISFSPTKDKKSSSNNQNLQAQISKEKTCETQR 310
V D LR ++ G+S+ F + D+ N+ +
Sbjct: 2715 VLNNDGLRVENLKCNTVGQSSGSFTDDEMGIDVDNDEVVELNHPTCSSLTGSSNGSSPMI 2894
Query: 311 NGSSSSSSSSSKGSWGKKMT 330
+GSSSSS S S GK T
Sbjct: 2895 HGSSSSSQSFENQSKGKSTT 2954
>TC11494 similar to UP|Q7X9B2 (Q7X9B2) 9/13 hydroperoxide lyase, partial
(27%)
Length = 541
Score = 31.6 bits (70), Expect = 0.24
Identities = 19/58 (32%), Positives = 26/58 (44%)
Frame = +2
Query: 29 DSNCSTPYVSAPSSPGRGGGPPPISSGYFYSAPASPLHFSITASSSYHHQTTTSSVPM 86
+S +TP S P+ PP + S +AP SP + T S S TT S P+
Sbjct: 260 ESKNTTPLSSEPTCLPAPSSPPTLESSRSSTAPLSPSSSTTTRSRSETSSTTLSCPPL 433
>TC18363 UP|Q56662 (Q56662) TcpI protein (Fragment), partial (9%)
Length = 594
Score = 31.2 bits (69), Expect = 0.32
Identities = 26/85 (30%), Positives = 38/85 (44%), Gaps = 10/85 (11%)
Frame = +1
Query: 68 SITASSSYHHQTTTSSVPMSYEFEFSARFGSTGSAASGS--MTSADELFLNGQIRPMKLS 125
S T + +H+Q T P +FEFS ++A+ +T AD L NGQI+P +
Sbjct: 130 SSTNQNKHHYQQVTKQDP---DFEFSTIKPDFNNSATSPFRITPADVLISNGQIKPHAIV 300
Query: 126 SHLER--------PQVLAPLLDLEE 142
R P L LL +E+
Sbjct: 301 CQQSRRPFVAKNAPMSLRSLLAVED 375
>TC13505 similar to UP|Q9T0K5 (Q9T0K5) Extensin-like protein, partial (7%)
Length = 359
Score = 30.8 bits (68), Expect = 0.42
Identities = 18/38 (47%), Positives = 20/38 (52%), Gaps = 3/38 (7%)
Frame = +1
Query: 30 SNCSTPYVSAPSSPGRGGGP---PPISSGYFYSAPASP 64
SNC P +P S G GGG PP S Y YS+P P
Sbjct: 214 SNCPPP--PSPPSGGGGGGSYYSPPPPSQYVYSSPPPP 321
>TC7794 homologue to UP|Q9M600 (Q9M600) Chlorophyll a/b binding protein
precursor, partial (97%)
Length = 2581
Score = 30.4 bits (67), Expect = 0.54
Identities = 23/85 (27%), Positives = 33/85 (38%), Gaps = 23/85 (27%)
Frame = -3
Query: 5 NSTKNPNLEPSNSNNGGET------------------FFEDIDSNCSTPYVSAPSSPGRG 46
N T +PNL P + + G+T F ++ + +P VS P SPG
Sbjct: 788 NHTASPNLTPLRAKSSGKTQPKAPNMAHLEWITSSSRFLANVSGSAESPAVSQP*SPGNS 609
Query: 47 -----GGPPPISSGYFYSAPASPLH 66
GG P +G Y + P H
Sbjct: 608 PVK*DGGSP--ENGPKYLTRSGPYH 540
>CB828239
Length = 552
Score = 30.4 bits (67), Expect = 0.54
Identities = 20/58 (34%), Positives = 30/58 (51%)
Frame = -2
Query: 237 PLDSASSSRSSSAGRSSKRWVFLKDFLRSKSEGRSNNKFWSTISFSPTKDKKSSSNNQ 294
P S SSS SSS+ S +++L F KS+ ++ +ST + T DK + NQ
Sbjct: 545 PSISPSSSSSSSSSSSYFLFLYLFFFFNPKSQKPPSSFLFSTTIINQTHDKIPNPQNQ 372
>BP050838
Length = 474
Score = 30.4 bits (67), Expect = 0.54
Identities = 17/62 (27%), Positives = 30/62 (47%)
Frame = +1
Query: 38 SAPSSPGRGGGPPPISSGYFYSAPASPLHFSITASSSYHHQTTTSSVPMSYEFEFSARFG 97
S P++ G PPP + + +++P S+ +S Y H + TS+ P + +A
Sbjct: 295 SMPAAVSGNGNPPPATLTFTFNSPT-----SMASSLCYSHPSPTSTNPTLFPSSTNAYSN 459
Query: 98 ST 99
ST
Sbjct: 460 ST 465
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.307 0.125 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,244,817
Number of Sequences: 28460
Number of extensions: 120287
Number of successful extensions: 1826
Number of sequences better than 10.0: 108
Number of HSP's better than 10.0 without gapping: 1244
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1579
length of query: 400
length of database: 4,897,600
effective HSP length: 92
effective length of query: 308
effective length of database: 2,279,280
effective search space: 702018240
effective search space used: 702018240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC149080.12


