

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149038.15 - phase: 0
(213 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
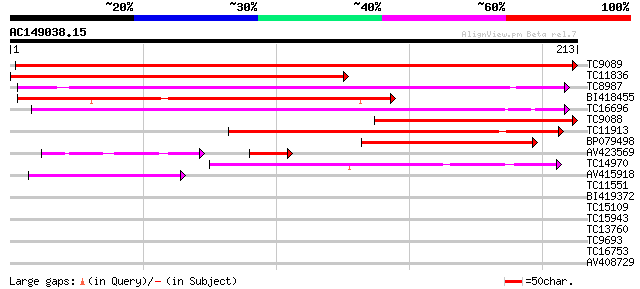
Score E
Sequences producing significant alignments: (bits) Value
TC9089 similar to UP|RHRE_PEA (Q9S8P4) Rhicadhesin receptor prec... 323 2e-89
TC11836 similar to GB|AAB51577.1|1755176|ATU75199 germin-like pr... 192 3e-50
TC8987 similar to UP|ABPA_PRUPE (Q9ZRA4) Auxin-binding protein A... 135 7e-33
BI418455 129 3e-31
TC16696 similar to UP|Q9AR81 (Q9AR81) Germin-like protein precur... 125 7e-30
TC9088 similar to UP|RHRE_PEA (Q9S8P4) Rhicadhesin receptor prec... 120 2e-28
TC11913 weakly similar to UP|ABPA_PRUPE (Q9ZRA4) Auxin-binding p... 91 2e-19
BP079498 62 6e-11
AV423569 36 8e-05
TC14970 similar to UP|O24294 (O24294) Legumin (Minor small) prec... 40 2e-04
AV415918 39 9e-04
TC11551 homologue to UP|AAQ93070 (AAQ93070) 3-ketoacyl-CoA thiol... 34 0.022
BI419372 27 2.0
TC15109 similar to UP|Q9FVE9 (Q9FVE9) Cytosolic aconitase, parti... 27 2.0
TC15943 similar to UP|Q8GT67 (Q8GT67) Xyloglucan-specific fungal... 27 2.7
TC13760 weakly similar to GB|AAM91086.1|22135799|AY128277 At1g45... 26 4.5
TC9693 similar to UP|COPP_MOUSE (O55029) Coatomer beta' subunit ... 26 5.9
TC16753 similar to UP|Q9SIA7 (Q9SIA7) Legumin-like protein, part... 25 7.8
AV408729 25 7.8
>TC9089 similar to UP|RHRE_PEA (Q9S8P4) Rhicadhesin receptor precursor
(Germin-like protein), partial (93%)
Length = 981
Score = 323 bits (827), Expect = 2e-89
Identities = 159/211 (75%), Positives = 183/211 (86%)
Frame = +1
Query: 3 IIAVLLLLVLTTTVTADPDNLQDLCVADLASAILVNGFTCKPASNVTAADFSTNVLAKPG 62
+ A+ L L+LTT ++DPD LQDLCVAD AS + VNGFTCK AS V A+DF +N+LAKPG
Sbjct: 178 LFALPLALLLTTAFSSDPDYLQDLCVADPASGVPVNGFTCKEASKVNASDFFSNILAKPG 357
Query: 63 ATNNTFGSLVTLANVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQLDV 122
ATNNTFGSLVT ANVQK+PGLNTLGVSLARIDYA GLNPPHTHPRATE+V+VLEG+LDV
Sbjct: 358 ATNNTFGSLVTGANVQKVPGLNTLGVSLARIDYAPDGLNPPHTHPRATEVVFVLEGELDV 537
Query: 123 GFITTANVLISKTIVKGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPLTLFA 182
GFITT NVLISK IVKG+ FVFPKGL+HFQKN G VPAAV++ FNSQL GT +I TLFA
Sbjct: 538 GFITTGNVLISKHIVKGDIFVFPKGLLHFQKNXGKVPAAVLSAFNSQLPGTQSIATTLFA 717
Query: 183 STPPVPDNILTQAFQIGTKEVQKIKSRLAPK 213
STP VPDN+LT+ FQ+GTK+++KIKSRLAPK
Sbjct: 718 STPTVPDNVLTKTFQVGTKQIEKIKSRLAPK 810
>TC11836 similar to GB|AAB51577.1|1755176|ATU75199 germin-like protein
{Arabidopsis thaliana;} , partial (56%)
Length = 434
Score = 192 bits (489), Expect = 3e-50
Identities = 93/127 (73%), Positives = 109/127 (85%)
Frame = +3
Query: 1 MKIIAVLLLLVLTTTVTADPDNLQDLCVADLASAILVNGFTCKPASNVTAADFSTNVLAK 60
MK++ +LLL + T T +DPD+LQD+CVADL+SAI VNGF CK AS V DF + +LAK
Sbjct: 54 MKLVVLLLLTLTTFTAASDPDSLQDICVADLSSAIKVNGFVCKSASAVKETDFFSAILAK 233
Query: 61 PGATNNTFGSLVTLANVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQL 120
PGATN TFGS+VT ANV+KIPGLNTLGVS++RIDYA GGLNPPHTHPRATE+V+VLEGQL
Sbjct: 234 PGATNTTFGSVVTGANVEKIPGLNTLGVSISRIDYAPGGLNPPHTHPRATEVVFVLEGQL 413
Query: 121 DVGFITT 127
DVGFITT
Sbjct: 414 DVGFITT 434
>TC8987 similar to UP|ABPA_PRUPE (Q9ZRA4) Auxin-binding protein ABP19a
precursor, partial (97%)
Length = 985
Score = 135 bits (339), Expect = 7e-33
Identities = 82/207 (39%), Positives = 117/207 (55%)
Frame = +1
Query: 4 IAVLLLLVLTTTVTADPDNLQDLCVADLASAILVNGFTCKPASNVTAADFSTNVLAKPGA 63
I L L+L+T+ A +QD CVADL + G+ CK + VT+ DF L+ PG
Sbjct: 37 IVFLFSLLLSTSHAA----VQDFCVADLKGSDGPAGYPCKTPATVTSDDFVFAGLSAPGN 204
Query: 64 TNNTFGSLVTLANVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQLDVG 123
N G+ VT A V + PG+N LG+S AR+D GG+ P HTHP A E++ V +G + G
Sbjct: 205 ITNIIGAAVTPAFVGQFPGVNGLGLSAARLDIGPGGVIPLHTHPGANELLIVTQGHITAG 384
Query: 124 FITTANVLISKTIVKGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPLTLFAS 183
FI++A+ + KT+ KGE VFP+GL+HFQ +G A A F+S G + LFAS
Sbjct: 385 FISSASTVYIKTLKKGELMVFPQGLLHFQVAAGKTKAIGFAIFSSASPGLQILDFALFAS 564
Query: 184 TPPVPDNILTQAFQIGTKEVQKIKSRL 210
P ++T+ + V+K+K L
Sbjct: 565 NFSTP--LITKTTFLDPALVKKLKGVL 639
>BI418455
Length = 477
Score = 129 bits (325), Expect = 3e-31
Identities = 74/149 (49%), Positives = 93/149 (61%), Gaps = 7/149 (4%)
Frame = +1
Query: 4 IAVLLLLVLTTTVTADPDNLQDLCVA--DLASAILVNGFTCKPASNVTAADFSTNVLAKP 61
+ +L L + DP LQD CV D + VNG CK + V A DF +V +P
Sbjct: 34 LVTILALASSFASAYDPSPLQDFCVGINDPKYGVFVNGKFCKDPALVKAEDFFKHV--EP 207
Query: 62 GATNNTFGSLVTLANVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQLD 121
G T+N GS VT V+++ GLNTLG+SLARID+A GLNPPHTHPRATEI+ VLEG L
Sbjct: 208 GNTSNPLGSKVTPVTVEQLFGLNTLGISLARIDFAPKGLNPPHTHPRATEILIVLEGTLY 387
Query: 122 VGFITTANV-----LISKTIVKGETFVFP 145
VGF+T+ N L +K + KG+ FVFP
Sbjct: 388 VGFVTSNNANGGNRLFTKVLNKGDVFVFP 474
>TC16696 similar to UP|Q9AR81 (Q9AR81) Germin-like protein precursor,
partial (96%)
Length = 834
Score = 125 bits (313), Expect = 7e-30
Identities = 75/202 (37%), Positives = 111/202 (54%)
Frame = +1
Query: 9 LLVLTTTVTADPDNLQDLCVADLASAILVNGFTCKPASNVTAADFSTNVLAKPGATNNTF 68
+L + + ++ ++ D CV DL G+ CK S VTA DF+ + L G T+
Sbjct: 7 ILFILSLLSLSHASVVDFCVGDLTFPNGPAGYACKKPSKVTADDFAYSGLGIAGNTSYII 186
Query: 69 GSLVTLANVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQLDVGFITTA 128
+ VT A + PG+N LG+SLAR+D A GG+ P HTHP A+E + V++G + GF+ +
Sbjct: 187 KAAVTPAFDAQFPGVNGLGISLARLDLASGGVIPLHTHPGASEALVVVQGTICAGFVASD 366
Query: 129 NVLISKTIVKGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPLTLFASTPPVP 188
N + KT+ KG+ VFP+GLVHFQ + G A F+S G + LF S P
Sbjct: 367 NTVYLKTLKKGDVMVFPQGLVHFQIHDGGSSALAFVSFSSANPGLQILDFALFKSDFP-S 543
Query: 189 DNILTQAFQIGTKEVQKIKSRL 210
D I AF + +++K+K L
Sbjct: 544 DLIAATAF-LDAAQIKKLKGVL 606
>TC9088 similar to UP|RHRE_PEA (Q9S8P4) Rhicadhesin receptor precursor
(Germin-like protein), partial (35%)
Length = 475
Score = 120 bits (301), Expect = 2e-28
Identities = 56/76 (73%), Positives = 67/76 (87%)
Frame = +3
Query: 138 KGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPLTLFASTPPVPDNILTQAFQ 197
KGE FVFPKGLVH+QKN+GY PA+VI+ FNSQL GTV+IPL LFA+ PPVPDN+LT+AFQ
Sbjct: 3 KGEIFVFPKGLVHYQKNNGYWPASVISAFNSQLPGTVSIPLALFAAKPPVPDNVLTKAFQ 182
Query: 198 IGTKEVQKIKSRLAPK 213
T+EV+KIK+ LAPK
Sbjct: 183 TSTEEVEKIKANLAPK 230
>TC11913 weakly similar to UP|ABPA_PRUPE (Q9ZRA4) Auxin-binding protein
ABP19a precursor, partial (61%)
Length = 530
Score = 90.9 bits (224), Expect = 2e-19
Identities = 45/126 (35%), Positives = 79/126 (61%)
Frame = +1
Query: 83 LNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQLDVGFITTANVLISKTIVKGETF 142
+N LG+S AR+D GG+ P H+HP A+E+V +L+G++ VGFI+T N + KT++KG+
Sbjct: 16 INGLGLSAARLDLDAGGVVPIHSHPGASELVIILQGRITVGFISTDNTVYQKTLMKGDII 195
Query: 143 VFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPLTLFASTPPVPDNILTQAFQIGTKE 202
V P+GL+HFQ N+G A+ + F+S + + LF + + ++ + + +
Sbjct: 196 VIPQGLLHFQLNAGGNRASAVLTFSSTNPVAQLVDVALFGNN--LDSGLVARTTFLDLAQ 369
Query: 203 VQKIKS 208
V+K+K+
Sbjct: 370 VKKLKA 387
>BP079498
Length = 373
Score = 62.4 bits (150), Expect = 6e-11
Identities = 29/66 (43%), Positives = 41/66 (61%)
Frame = -2
Query: 133 SKTIVKGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPLTLFASTPPVPDNIL 192
+K + KG+ FVFP L+HFQ N G A IAG +SQ G + I LF ++PP+ +L
Sbjct: 372 TKVLNKGDVFVFPISLIHFQLNVGSGNAVAIAGLSSQNPGVITIANALFKASPPISQEVL 193
Query: 193 TQAFQI 198
T+A Q+
Sbjct: 192 TKALQV 175
>AV423569
Length = 351
Score = 36.2 bits (82), Expect(2) = 8e-05
Identities = 14/16 (87%), Positives = 15/16 (93%)
Frame = +3
Query: 91 ARIDYAVGGLNPPHTH 106
+RIDYA GGLNPPHTH
Sbjct: 303 SRIDYAPGGLNPPHTH 350
Score = 25.0 bits (53), Expect(2) = 8e-05
Identities = 20/61 (32%), Positives = 30/61 (48%)
Frame = +2
Query: 13 TTTVTADPDNLQDLCVADLASAILVNGFTCKPASNVTAADFSTNVLAKPGATNNTFGSLV 72
T T +DPD L ++CV + +P+ T + T+ + ATN TFGS+V
Sbjct: 101 TFTAASDPD-LYNICVHYPPXRM----DRLQPSDKETDSSHHTS---QTSATNTTFGSVV 256
Query: 73 T 73
T
Sbjct: 257 T 259
>TC14970 similar to UP|O24294 (O24294) Legumin (Minor small) precursor,
partial (51%)
Length = 1566
Score = 40.4 bits (93), Expect = 2e-04
Identities = 32/133 (24%), Positives = 60/133 (45%), Gaps = 1/133 (0%)
Frame = +1
Query: 76 NVQKIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQLDVGFIT-TANVLISK 134
N +P L LG+S ++ G+ PH + A I+YV+ G+ V + + +
Sbjct: 937 NSLTLPILRFLGLSAEYVNLYQNGIYGPHWNINANSIIYVVRGRGRVRIVNCQGQAVFND 1116
Query: 135 TIVKGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPLTLFASTPPVPDNILTQ 194
+ KG+ V P+ V Q+ V+ F + + V+ +F +TP +L+
Sbjct: 1117 ELRKGQLLVVPQNFVVAQQAQDEGFEYVV--FKTNARAAVSHVKQVFRATPA---EVLSN 1281
Query: 195 AFQIGTKEVQKIK 207
AF I +++ +K
Sbjct: 1282 AFGIRQRDISDLK 1320
>AV415918
Length = 189
Score = 38.5 bits (88), Expect = 9e-04
Identities = 19/59 (32%), Positives = 30/59 (50%)
Frame = +3
Query: 8 LLLVLTTTVTADPDNLQDLCVADLASAILVNGFTCKPASNVTAADFSTNVLAKPGATNN 66
+L + T T ++ DLCVAD +G+ C P +N+T+ DF + L TN+
Sbjct: 6 ILFLFTFLSTTSYASVNDLCVADFKGPSTPSGYHCMPLANLTSDDFVFHGLVAGNTTNS 182
>TC11551 homologue to UP|AAQ93070 (AAQ93070) 3-ketoacyl-CoA thiolase ,
partial (37%)
Length = 915
Score = 33.9 bits (76), Expect = 0.022
Identities = 25/72 (34%), Positives = 39/72 (53%)
Frame = -2
Query: 14 TTVTADPDNLQDLCVADLASAILVNGFTCKPASNVTAADFSTNVLAKPGATNNTFGSLVT 73
T + + P+ LQ+ D A+A L++ + P+S ADF+ +AK G T T GS T
Sbjct: 320 TFLVSSPNFLQE*TNCD-ANASLISNKSMLPSSR--PADFTAAGIAKAGPTPMTEGSTPT 150
Query: 74 LANVQKIPGLNT 85
A + KIP + +
Sbjct: 149 AAKLLKIPRIGS 114
>BI419372
Length = 564
Score = 27.3 bits (59), Expect = 2.0
Identities = 18/50 (36%), Positives = 25/50 (50%), Gaps = 9/50 (18%)
Frame = +3
Query: 62 GATNNTFGSLVTLANVQKIPGLNTLGVSLARIDY---------AVGGLNP 102
GAT++ GSL T A + + ++ +G L IDY A GGL P
Sbjct: 363 GATDSWHGSLATAALIGFMGPISAIGAILTGIDYSGLDHIMVFACGGLIP 512
>TC15109 similar to UP|Q9FVE9 (Q9FVE9) Cytosolic aconitase, partial (46%)
Length = 1237
Score = 27.3 bits (59), Expect = 2.0
Identities = 20/57 (35%), Positives = 27/57 (47%)
Frame = -1
Query: 32 ASAILVNGFTCKPASNVTAADFSTNVLAKPGATNNTFGSLVTLANVQKIPGLNTLGV 88
A +L++GFTC P S+ A F+TN F LV +A + P L GV
Sbjct: 175 ARLVLIHGFTCNPNSH---AFFATNAAPSITLGFEVFVQLV-IAAITTSPCLRCAGV 17
>TC15943 similar to UP|Q8GT67 (Q8GT67) Xyloglucan-specific fungal
endoglucanase inhibitor protein precursor, partial (15%)
Length = 968
Score = 26.9 bits (58), Expect = 2.7
Identities = 18/37 (48%), Positives = 20/37 (53%), Gaps = 2/37 (5%)
Frame = +2
Query: 43 KPASNVTAADFSTNVLAKPGATNNTFGS--LVTLANV 77
K SN A S + KPG TNNT G+ L LANV
Sbjct: 329 KSCSNTGAGCTSCDGPLKPGCTNNTCGANILNPLANV 439
>TC13760 weakly similar to GB|AAM91086.1|22135799|AY128277 At1g45261/F2G19.1
{Arabidopsis thaliana;}, partial (18%)
Length = 403
Score = 26.2 bits (56), Expect = 4.5
Identities = 20/64 (31%), Positives = 28/64 (43%)
Frame = -2
Query: 123 GFITTANVLISKTIVKGETFVFPKGLVHFQKNSGYVPAAVIAGFNSQLQGTVNIPLTLFA 182
GF TT+N++ +V E + L+H Q S +VP Q G N+P L
Sbjct: 339 GFKTTSNLIFRIWMVSKELVYYSFLLLHIQYLSIHVPWREKNVLGKQNDG-FNVPENLLF 163
Query: 183 STPP 186
PP
Sbjct: 162 IIPP 151
>TC9693 similar to UP|COPP_MOUSE (O55029) Coatomer beta' subunit
(Beta'-coat protein) (Beta'-COP) (p102), partial (20%)
Length = 743
Score = 25.8 bits (55), Expect = 5.9
Identities = 12/27 (44%), Positives = 16/27 (58%)
Frame = +3
Query: 20 PDNLQDLCVADLASAILVNGFTCKPAS 46
P +LQ CV+ L S I+ + CKP S
Sbjct: 9 PPSLQRRCVSTLGSIIIFPLWLCKPRS 89
>TC16753 similar to UP|Q9SIA7 (Q9SIA7) Legumin-like protein, partial (42%)
Length = 534
Score = 25.4 bits (54), Expect = 7.8
Identities = 17/76 (22%), Positives = 34/76 (44%)
Frame = +1
Query: 79 KIPGLNTLGVSLARIDYAVGGLNPPHTHPRATEIVYVLEGQLDVGFITTANVLISKTIVK 138
++P L + +++ G P + ++++ YVL+G VG + + I K
Sbjct: 160 ELPMLREGNIGASKLALQKNGFALPR-YSDSSKVAYVLQGTGVVGIVLPESEEKVVAIKK 336
Query: 139 GETFVFPKGLVHFQKN 154
G+ P G+V + N
Sbjct: 337 GDALALPLGVVTWWYN 384
>AV408729
Length = 433
Score = 25.4 bits (54), Expect = 7.8
Identities = 13/29 (44%), Positives = 17/29 (57%)
Frame = +3
Query: 154 NSGYVPAAVIAGFNSQLQGTVNIPLTLFA 182
+ G+VP +IAGF + T NI L L A
Sbjct: 144 DQGWVPIKLIAGFKKVMHLTDNIQLILDA 230
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.136 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,123,375
Number of Sequences: 28460
Number of extensions: 35678
Number of successful extensions: 206
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 202
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 203
length of query: 213
length of database: 4,897,600
effective HSP length: 86
effective length of query: 127
effective length of database: 2,450,040
effective search space: 311155080
effective search space used: 311155080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 53 (25.0 bits)
Medicago: description of AC149038.15


