

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148995.5 + phase: 0
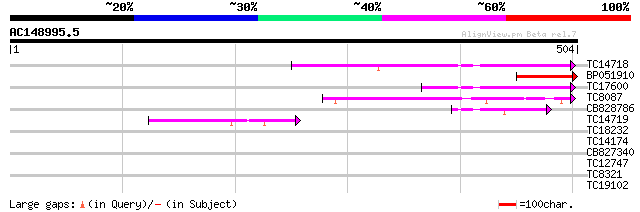
(504 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14718 similar to UP|NCPR_CATRO (Q05001) NADPH--cytochrome P450... 174 4e-44
BP051910 99 2e-21
TC17600 similar to PIR|A47298|A47298 NADPH-ferrihemoprotein redu... 90 1e-18
TC8087 homologue to UP|FENR_PEA (P10933) Ferredoxin--NADP reduct... 58 4e-09
CB828786 54 6e-08
TC14719 similar to UP|Q9AU07 (Q9AU07) NADPH-cytochrome P450 oxyd... 53 1e-07
TC18232 similar to UP|Q7PS48 (Q7PS48) ENSANGP00000015589 (Fragme... 29 1.6
TC14174 UP|PSBC_LOTJA (Q9BBT1) Photosystem II 44 kDa reaction ce... 28 2.7
CB827340 28 4.6
TC12747 weakly similar to UP|Q8LAH2 (Q8LAH2) Polygalacturonase-l... 28 4.6
TC8321 similar to UP|AAS46230 (AAS46230) Peroxiredoxin Q, partia... 23 7.8
TC19102 similar to UP|Q40484 (Q40484) Cdc2 homolog, partial (28%) 27 7.8
>TC14718 similar to UP|NCPR_CATRO (Q05001) NADPH--cytochrome P450 reductase
(CPR) (P450R) , partial (36%)
Length = 1163
Score = 174 bits (440), Expect = 4e-44
Identities = 92/260 (35%), Positives = 154/260 (58%), Gaps = 7/260 (2%)
Frame = +1
Query: 251 RRTVLEVLKDFPSVQMPLE-WLIQLVPMLKKREFSISSSQSSHPNQVHLTVSVVSWTTPY 309
+R++LEV+ +FPS + P+ + + P L+ R +SISSS P+++H+T ++V+ P
Sbjct: 16 QRSLLEVMAEFPSAKPPIGVFFAAIAPRLQPRFYSISSSPRMAPSRIHVTCALVNDKMPT 195
Query: 310 KRKKKGLCSSWLAALDP----RDAVSLPVWFQKGSLPTPSPS-LPLILVGPGTGCAPFRG 364
R +G+CS+W+ P +D P++ ++ + P+ + +P+I++GPGTG APFRG
Sbjct: 196 GRIHRGVCSTWMKNSVPLEKSQDCSWAPIFVRQSNFKLPADNKVPIIMIGPGTGLAPFRG 375
Query: 365 FIEER-ALQSKTISIAPIIFFFGCWNEDGDFLYKDFWLNHSQNNGVLSESTGGGFYVAFS 423
F++ER AL+ + P + FFGC N D++Y+D LNH N+G LSE VAFS
Sbjct: 376 FLQERLALKEDGAELGPSVLFFGCRNRQMDYIYEDE-LNHFVNSGALSE-----LIVAFS 537
Query: 424 RDQPEKVYVQHKLREHSGRVWNLLAEGASVYIAGSLTKMPTDVTSAFEEIVSKENDVSKE 483
R+ P K YVQHK+ E + +WN++++GA +Y+ G M DV I+ ++ +
Sbjct: 538 REGPTKEYVQHKMMEKASDIWNMISQGAYIYVCGDAKGMARDVHRTLHTILQEQGSLDSS 717
Query: 484 DAVRWIRALEKCGKYHIEAW 503
A ++ L+ G+Y + W
Sbjct: 718 KAEGMVKNLQLNGRYLRDVW 777
>BP051910
Length = 429
Score = 99.0 bits (245), Expect = 2e-21
Identities = 46/54 (85%), Positives = 51/54 (94%)
Frame = -3
Query: 451 ASVYIAGSLTKMPTDVTSAFEEIVSKENDVSKEDAVRWIRALEKCGKYHIEAWS 504
A+VYIAGS T+MP DVTSAFEEI+SKEN+VSKEDAVRWIRALEK GKYH+EAWS
Sbjct: 427 AAVYIAGSSTQMPADVTSAFEEIISKENEVSKEDAVRWIRALEKSGKYHVEAWS 266
>TC17600 similar to PIR|A47298|A47298 NADPH-ferrihemoprotein reductase -
mung bean {Vigna radiata;} , partial (19%)
Length = 717
Score = 89.7 bits (221), Expect = 1e-18
Identities = 50/137 (36%), Positives = 74/137 (53%)
Frame = +3
Query: 367 EERALQSKTISIAPIIFFFGCWNEDGDFLYKDFWLNHSQNNGVLSESTGGGFYVAFSRDQ 426
E AL+ I + P I FFGC N DF+Y+D LN+ G LSE VAFSR+
Sbjct: 6 ERFALKEDGIELGPAILFFGCRNRRMDFIYEDE-LNNFLEQGSLSE-----LIVAFSREG 167
Query: 427 PEKVYVQHKLREHSGRVWNLLAEGASVYIAGSLTKMPTDVTSAFEEIVSKENDVSKEDAV 486
EK YVQHK+ + + +W+L+++GA +Y+ G M DV IV ++ +V A
Sbjct: 168 SEKEYVQHKMMDKAAYLWSLISQGAYLYVCGDAKGMARDVHHTLHTIVQQQENVESSKAE 347
Query: 487 RWIRALEKCGKYHIEAW 503
++ L+ G+Y + W
Sbjct: 348 AIVKKLQLDGRYLRDVW 398
>TC8087 homologue to UP|FENR_PEA (P10933) Ferredoxin--NADP reductase, leaf
isozyme, chloroplast precursor (FNR) , partial (94%)
Length = 1449
Score = 57.8 bits (138), Expect = 4e-09
Identities = 57/239 (23%), Positives = 103/239 (42%), Gaps = 14/239 (5%)
Frame = +1
Query: 279 KKREFSISSS---QSSHPNQVHLTVSVVSWTTPYKRKKKGLCSSWLAALDPRDAVSLPVW 335
K R +SI+SS V L V + +T KG+CS++L L P V++
Sbjct: 553 KLRLYSIASSALGDFGDSKTVSLCVKRLVYTNDSGEIVKGVCSNFLCDLKPGSEVTITGP 732
Query: 336 FQKGSLPTPSPSLPLILVGPGTGCAPFRGFIEERALQS-KTISIAPIIFFFGCWNEDGDF 394
K L P+ +I++ GTG APFR F+ + + + + F
Sbjct: 733 VGKEMLMPKDPNATVIMLATGTGIAPFRSFLWKMFFEKHDDYKFNGLAWLFLGVPTSSSL 912
Query: 395 LYKDFWLNHSQNNGVLSESTGGGFYVAF------SRDQPEKVYVQHKLREHSGRVWNLL- 447
LYK+ + + E + F + F + D+ EK+Y+Q ++ +++ +W LL
Sbjct: 913 LYKEEFEK-------MKEKSPENFRLDFAVSREQTNDKGEKMYIQTRMAQYAEELWQLLK 1071
Query: 448 AEGASVYIAGSLTKMPTDVTSAFEEIVSKENDVSKEDAVRWI---RALEKCGKYHIEAW 503
+ VY+ G L M + + ++ D + WI R L+K ++++E +
Sbjct: 1072KDNTFVYMCG-LKGMEKGIDDIMVSLAAR-------DGIDWIEYKRQLKKAEQWNVEVY 1224
>CB828786
Length = 456
Score = 53.9 bits (128), Expect = 6e-08
Identities = 35/117 (29%), Positives = 53/117 (44%), Gaps = 28/117 (23%)
Frame = +1
Query: 393 DFLYKDFWLNHSQNNGVLSESTGGGFYVAFSRDQPEKVYVQHKLRE-------------- 438
D++Y+D LNH N+G LSE VAFSR+ P K YVQHK+ E
Sbjct: 4 DYIYEDE-LNHFVNSGALSE-----LIVAFSREGPTKEYVQHKMMEKVPILSQDAFNYQF 165
Query: 439 --------------HSGRVWNLLAEGASVYIAGSLTKMPTDVTSAFEEIVSKENDVS 481
+ +WN++++GA +Y+ G M DV I+ ++ +S
Sbjct: 166 TIYSFSY*TLYLCGQASDIWNMISQGAYIYVCGDAKGMARDVHRTLHTILQEQVYIS 336
>TC14719 similar to UP|Q9AU07 (Q9AU07) NADPH-cytochrome P450 oxydoreductase
isoform 2, partial (28%)
Length = 633
Score = 53.1 bits (126), Expect = 1e-07
Identities = 43/145 (29%), Positives = 67/145 (45%), Gaps = 10/145 (6%)
Frame = +3
Query: 124 LTKPNSGKDVRHFEFEFVSHAIEYDTGDILEVLPGQDSAAVDAFIRRCNLDPDSLITVSP 183
L P S + H EF+ + Y+TGD + V S V+ +R L PD+ +V
Sbjct: 201 LHTPVSDRSCTHLEFDISGTGVAYETGDHVGVYCENLSETVEEAVRLLGLSPDTYFSVHT 380
Query: 184 KGMDCNG-HGSRM-----PVKLRTFVELTMDVASASPRRYFFEARCSE----HERERLEY 233
D GS + P LRT + DV S SP++ A + E +RL +
Sbjct: 381 DDEDGKPLSGSSLPPTFPPCTLRTAIARYADVLS-SPKKSGLLALAAHASNPSEADRLRH 557
Query: 234 FASPEGRDDLYQYNQKERRTVLEVL 258
ASP G+D+ ++ +R++LEV+
Sbjct: 558 LASPAGKDEYSEWVIASQRSLLEVM 632
>TC18232 similar to UP|Q7PS48 (Q7PS48) ENSANGP00000015589 (Fragment),
partial (8%)
Length = 907
Score = 29.3 bits (64), Expect = 1.6
Identities = 11/28 (39%), Positives = 17/28 (60%)
Frame = -3
Query: 392 GDFLYKDFWLNHSQNNGVLSESTGGGFY 419
G ++K W NHS++ VL+ GGG +
Sbjct: 395 GSVVHK*QWGNHSEHRNVLARQAGGGIF 312
>TC14174 UP|PSBC_LOTJA (Q9BBT1) Photosystem II 44 kDa reaction center protein
(P6 protein) (CP43), complete
Length = 3406
Score = 28.5 bits (62), Expect = 2.7
Identities = 14/28 (50%), Positives = 19/28 (67%)
Frame = -2
Query: 59 SGPDVLIQDTTLIDQPKVQITYHNIENV 86
SGPD L+Q+ + +Q VQIT N+ NV
Sbjct: 1380 SGPDRLLQEHSSTNQLLVQIT*RNVTNV 1297
>CB827340
Length = 555
Score = 27.7 bits (60), Expect = 4.6
Identities = 19/57 (33%), Positives = 26/57 (45%)
Frame = -2
Query: 236 SPEGRDDLYQYNQKERRTVLEVLKDFPSVQMPLEWLIQLVPMLKKREFSISSSQSSH 292
SP G+DD +Y VL +LK+ ++P L P L +SSSQ H
Sbjct: 185 SPSGKDDPQKYQNMLNILVLGILKN--PYELPHSISCPLEPFLPCLSRCLSSSQDLH 21
>TC12747 weakly similar to UP|Q8LAH2 (Q8LAH2) Polygalacturonase-like
protein, partial (43%)
Length = 624
Score = 27.7 bits (60), Expect = 4.6
Identities = 19/71 (26%), Positives = 33/71 (45%), Gaps = 12/71 (16%)
Frame = +2
Query: 32 SGYEGTLDPWLSSLWRM-----LNMIKPEL--LPSGPDVLIQDTTLIDQPK-----VQIT 79
+GY+GT+D S W + LN +P + L ++ I + T+++ P V
Sbjct: 98 TGYDGTIDGQGSVWWDLISTGDLNYSRPHIIELIGSDNITISNLTILNSPSWGIHPVYCR 277
Query: 80 YHNIENVVSHS 90
Y I N+ H+
Sbjct: 278 YVQIHNITVHA 310
>TC8321 similar to UP|AAS46230 (AAS46230) Peroxiredoxin Q, partial (75%)
Length = 873
Score = 23.1 bits (48), Expect(2) = 7.8
Identities = 9/23 (39%), Positives = 14/23 (60%)
Frame = +1
Query: 340 SLPTPSPSLPLILVGPGTGCAPF 362
++P P+ SLP+ L P + PF
Sbjct: 16 TVPNPTHSLPIFLHPPSSSNLPF 84
Score = 21.9 bits (45), Expect(2) = 7.8
Identities = 11/26 (42%), Positives = 14/26 (53%)
Frame = +3
Query: 376 ISIAPIIFFFGCWNEDGDFLYKDFWL 401
ISI PIIF F + F + F+L
Sbjct: 81 ISIPPIIFQFSIYWPQAIFFFFSFFL 158
>TC19102 similar to UP|Q40484 (Q40484) Cdc2 homolog, partial (28%)
Length = 639
Score = 26.9 bits (58), Expect = 7.8
Identities = 13/42 (30%), Positives = 21/42 (49%)
Frame = +3
Query: 139 EFVSHAIEYDTGDILEVLPGQDSAAVDAFIRRCNLDPDSLIT 180
+F S ++ + D+ V+P DSA +D LDP +T
Sbjct: 96 DFKSAFPKWPSKDLATVVPNLDSAGLDLLSNMLRLDPTKRVT 221
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.136 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,756,940
Number of Sequences: 28460
Number of extensions: 147346
Number of successful extensions: 801
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 790
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 794
length of query: 504
length of database: 4,897,600
effective HSP length: 94
effective length of query: 410
effective length of database: 2,222,360
effective search space: 911167600
effective search space used: 911167600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC148995.5


