

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148994.1 + phase: 0
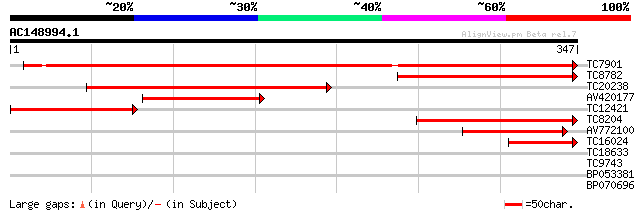
(347 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC7901 homologue to UP|UPT2_SOLTU (Q8RU27) Alpha-1,4-glucan-prot... 366 e-102
TC8782 weakly similar to GB|AAM66046.1|21594813|AY088511 amyloge... 206 5e-54
TC20238 homologue to UP|O22427 (O22427) Reversibly glycosylated ... 187 2e-48
AV420177 153 4e-38
TC12421 weakly similar to GB|AAM66046.1|21594813|AY088511 amylog... 131 2e-31
TC8204 homologue to UP|O24547 (O24547) Type IIIa membrane protei... 115 8e-27
AV772100 59 1e-09
TC16024 similar to UP|O22428 (O22428) Reversibly glycosylated po... 50 7e-07
TC18633 similar to GB|AAB94051.1|2736075|AF016644 PAP-specific p... 27 3.9
TC9743 similar to UP|AAQ89667 (AAQ89667) At5g25170, partial (36%) 27 5.1
BP053381 26 8.6
BP070696 26 8.6
>TC7901 homologue to UP|UPT2_SOLTU (Q8RU27) Alpha-1,4-glucan-protein
synthase [UDP-forming] 2 (UDP-glucose:protein
transglucosylase 2) (UPTG 2) , partial (92%)
Length = 1451
Score = 366 bits (939), Expect = e-102
Identities = 169/340 (49%), Positives = 240/340 (69%), Gaps = 1/340 (0%)
Frame = +1
Query: 9 NEVDIVIGALHSNLTPFMNEWKSIFSRFHLIIVKDPALKEELQIPEGFSADVYTNSEIER 68
+E+DIVI + + F+ W+ F +HLIIV+D + + +PEGF ++Y ++I R
Sbjct: 97 DELDIVIPTIRN--LDFLEMWRPFFQGYHLIIVQDGDPSKTINVPEGFDYELYNRNDINR 270
Query: 69 VVGSSTS-IRFSGYACRYFGFLVSKKKYVVCIDDDCVPAKDDAGNVVDAVAQHIVNLKTP 127
++G S I F ACR FG++VSKKKYV IDDDC AKD +G ++A+ QHI NL TP
Sbjct: 271 ILGPKASCISFKDSACRCFGYMVSKKKYVYTIDDDCFVAKDPSGKDINALEQHIKNLLTP 450
Query: 128 ATPFFFNTLYDPFRKGADFVRGYPFSLRSGVDCALSCGLWLNLADLDAPTQALKPTQRNS 187
+TP FFNTLYDP+R+GADFVRGYPFSLR G A+S GLWLN+ D DAPTQ +KP +RN+
Sbjct: 451 STPHFFNTLYDPYREGADFVRGYPFSLREGAPTAVSHGLWLNIPDYDAPTQLVKPRERNT 630
Query: 188 RYVDAVLTVPTRAMLPVSGINIAFNRELVGPALVPALVLAGEGKLRWETVEDIWCGLCVK 247
RYVDAV+T+P + P+ G+N+AF+R+L+GPA+ L+ G+ R+ +D+W G C+K
Sbjct: 631 RYVDAVMTIPKGTLFPMCGMNLAFDRQLIGPAMYFGLMGDGQPIGRY---DDMWAGWCIK 801
Query: 248 IVCDHLSLGVKSGLPYVWRNERGNAIDSLKKEWEGVKLMEDVVPFFQSVKLPQSATTAED 307
++CDHL LGVK+GLPY+W ++ N +LKKE++G+ E+++PFFQS LP+ T+ +
Sbjct: 802 VICDHLGLGVKTGLPYIWHSKASNPFVNLKKEYKGIFWQEEIIPFFQSATLPKDCTSVQK 981
Query: 308 CVIEMAKSVKEQLGKVDPMFLKAADAMAEWVKLWKSVGSA 347
C IE++K VKE+LG VDP F K DAM W++ W + ++
Sbjct: 982 CYIELSKQVKEKLGPVDPYFNKLGDAMVTWIEAWDELNNS 1101
>TC8782 weakly similar to GB|AAM66046.1|21594813|AY088511 amylogenin
{Arabidopsis thaliana;}, partial (31%)
Length = 727
Score = 206 bits (524), Expect = 5e-54
Identities = 94/110 (85%), Positives = 103/110 (93%)
Frame = +1
Query: 238 EDIWCGLCVKIVCDHLSLGVKSGLPYVWRNERGNAIDSLKKEWEGVKLMEDVVPFFQSVK 297
EDIWCG+CVK++CDHL LGVKSGLPYVWR ERG+AI+SLKKEWEGVKLMEDVVPFFQSV+
Sbjct: 1 EDIWCGMCVKVICDHLGLGVKSGLPYVWRTERGDAIESLKKEWEGVKLMEDVVPFFQSVR 180
Query: 298 LPQSATTAEDCVIEMAKSVKEQLGKVDPMFLKAADAMAEWVKLWKSVGSA 347
LPQSATTAEDCV+EMAK+VKE LGKVDPMF AA+ M EWVKLWKSVGSA
Sbjct: 181 LPQSATTAEDCVVEMAKTVKENLGKVDPMFSHAAETMEEWVKLWKSVGSA 330
>TC20238 homologue to UP|O22427 (O22427) Reversibly glycosylated
polypeptide-1, partial (43%)
Length = 466
Score = 187 bits (475), Expect = 2e-48
Identities = 84/151 (55%), Positives = 113/151 (74%), Gaps = 1/151 (0%)
Frame = +1
Query: 48 EELQIPEGFSADVYTNSEIERVVGSSTS-IRFSGYACRYFGFLVSKKKYVVCIDDDCVPA 106
+ + +P+GF ++Y ++I R++G + I F ACR FG++VSKKKY+ IDDDC A
Sbjct: 1 QTIHVPDGFDYELYNRNDINRILGPKANCISFKDSACRCFGYMVSKKKYIYTIDDDCFVA 180
Query: 107 KDDAGNVVDAVAQHIVNLKTPATPFFFNTLYDPFRKGADFVRGYPFSLRSGVDCALSCGL 166
D +G ++A+ QHI NL P+TP+FFNTLY+P+R+GADFVRGYPFSLR GV A S GL
Sbjct: 181 NDPSGKKINALEQHIKNLLCPSTPYFFNTLYEPYREGADFVRGYPFSLREGVPTAASHGL 360
Query: 167 WLNLADLDAPTQALKPTQRNSRYVDAVLTVP 197
WLN+ D DAPTQ +KP +RN+RYVD V+T+P
Sbjct: 361 WLNIPDYDAPTQLVKPLERNTRYVDMVMTIP 453
>AV420177
Length = 227
Score = 153 bits (387), Expect = 4e-38
Identities = 69/75 (92%), Positives = 74/75 (98%)
Frame = +2
Query: 82 ACRYFGFLVSKKKYVVCIDDDCVPAKDDAGNVVDAVAQHIVNLKTPATPFFFNTLYDPFR 141
+CRYFG L+S+KKYVVCIDDDCVPAKDD+GN+VDAVAQHIVNLKTPATPFFFNTLYDPFR
Sbjct: 2 SCRYFGILISRKKYVVCIDDDCVPAKDDSGNLVDAVAQHIVNLKTPATPFFFNTLYDPFR 181
Query: 142 KGADFVRGYPFSLRS 156
KGADFVRGYPFSLRS
Sbjct: 182 KGADFVRGYPFSLRS 226
>TC12421 weakly similar to GB|AAM66046.1|21594813|AY088511 amylogenin
{Arabidopsis thaliana;}, partial (22%)
Length = 466
Score = 131 bits (329), Expect = 2e-31
Identities = 63/78 (80%), Positives = 70/78 (88%)
Frame = +1
Query: 1 MSQVIINDNEVDIVIGALHSNLTPFMNEWKSIFSRFHLIIVKDPALKEELQIPEGFSADV 60
MSQ IINDNEVDIVIGALH +LT FMNEW+ +FSRFHLIIVKDP L EELQIPEGFSADV
Sbjct: 232 MSQAIINDNEVDIVIGALHPDLTSFMNEWRPVFSRFHLIIVKDPDLNEELQIPEGFSADV 411
Query: 61 YTNSEIERVVGSSTSIRF 78
YT S+I+RV GSS+S+RF
Sbjct: 412 YTKSDIDRVAGSSSSLRF 465
>TC8204 homologue to UP|O24547 (O24547) Type IIIa membrane protein
cp-wap11, partial (34%)
Length = 777
Score = 115 bits (289), Expect = 8e-27
Identities = 49/98 (50%), Positives = 70/98 (71%)
Frame = +3
Query: 250 CDHLSLGVKSGLPYVWRNERGNAIDSLKKEWEGVKLMEDVVPFFQSVKLPQSATTAEDCV 309
CDHL LG+K+GLPY++ ++ N +L+KE++G+ ED++PFFQS LP+ ATT + C
Sbjct: 3 CDHLGLGIKTGLPYIYHSKASNPFVNLRKEYKGIFWQEDIIPFFQSAVLPKEATTVQKCY 182
Query: 310 IEMAKSVKEQLGKVDPMFLKAADAMAEWVKLWKSVGSA 347
IE+AK VKE+L K+DP F K ADAM W++ W + A
Sbjct: 183 IELAKQVKEKLTKIDPYFDKLADAMVTWIEAWDELNPA 296
>AV772100
Length = 435
Score = 58.9 bits (141), Expect = 1e-09
Identities = 23/65 (35%), Positives = 40/65 (61%), Gaps = 1/65 (1%)
Frame = -2
Query: 278 KEWEGVKLMEDVVPFFQSVKLPQSATTAEDCVIEMAKSV-KEQLGKVDPMFLKAADAMAE 336
KE++G+ +E+++PFF S LP+ T+ C +E+++ + L VDP F + DAM
Sbjct: 434 KEYQGIFCLEEIIPFFPSATLPKDCTSVPKCYLELSQPTSRRSLALVDPYFYQLGDAMVT 255
Query: 337 WVKLW 341
W++ W
Sbjct: 254 WIEAW 240
>TC16024 similar to UP|O22428 (O22428) Reversibly glycosylated
polypeptide-2, partial (12%)
Length = 544
Score = 49.7 bits (117), Expect = 7e-07
Identities = 20/42 (47%), Positives = 28/42 (66%)
Frame = +2
Query: 306 EDCVIEMAKSVKEQLGKVDPMFLKAADAMAEWVKLWKSVGSA 347
+ C IE++K VKE+L K+DP F K ADAM W++ W + A
Sbjct: 2 QKCYIELSKQVKEKLSKIDPYFDKLADAMVTWIEAWDQLNPA 127
>TC18633 similar to GB|AAB94051.1|2736075|AF016644 PAP-specific phosphatase
{Arabidopsis thaliana;} , partial (45%)
Length = 882
Score = 27.3 bits (59), Expect = 3.9
Identities = 15/38 (39%), Positives = 19/38 (49%)
Frame = +3
Query: 5 IINDNEVDIVIGALHSNLTPFMNEWKSIFSRFHLIIVK 42
+I D EV V+G L P EW S R+H I+ K
Sbjct: 543 LIEDGEV--VLGVLGCPNYPMRKEWLSYQHRYHRIVAK 650
>TC9743 similar to UP|AAQ89667 (AAQ89667) At5g25170, partial (36%)
Length = 552
Score = 26.9 bits (58), Expect = 5.1
Identities = 19/65 (29%), Positives = 33/65 (50%), Gaps = 8/65 (12%)
Frame = -3
Query: 215 LVGPALVPALVLAGE-GKLR------WETVEDIWCGLCVKIV-CDHLSLGVKSGLPYVWR 266
LVG ++ +V+ G+ GK R WE + +W LC+K++ H ++ LP+ +
Sbjct: 214 LVGWLVMALVVVMGQNGKSREIPRAYWEPRKTLWEILCLKVIPGSHYNM*CTLSLPFAFG 35
Query: 267 NERGN 271
R N
Sbjct: 34 ERRRN 20
>BP053381
Length = 524
Score = 26.2 bits (56), Expect = 8.6
Identities = 14/45 (31%), Positives = 19/45 (42%)
Frame = +1
Query: 39 IIVKDPALKEELQIPEGFSADVYTNSEIERVVGSSTSIRFSGYAC 83
I+ P + LQI GFS D + SS + S Y+C
Sbjct: 256 ILPSSPQILRSLQIFRGFSVDCQQEPSYKHT*FSSQNSSLSSYSC 390
>BP070696
Length = 429
Score = 26.2 bits (56), Expect = 8.6
Identities = 13/36 (36%), Positives = 20/36 (55%)
Frame = -3
Query: 277 KKEWEGVKLMEDVVPFFQSVKLPQSATTAEDCVIEM 312
KKEW+G+ E++ F LP + TT D +I +
Sbjct: 316 KKEWKGI**SEEMEISFVLCYLPLAKTTLSDFLIRL 209
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.137 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,125,570
Number of Sequences: 28460
Number of extensions: 82803
Number of successful extensions: 372
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 369
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 370
length of query: 347
length of database: 4,897,600
effective HSP length: 91
effective length of query: 256
effective length of database: 2,307,740
effective search space: 590781440
effective search space used: 590781440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC148994.1


