

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148970.6 - phase: 0
(355 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
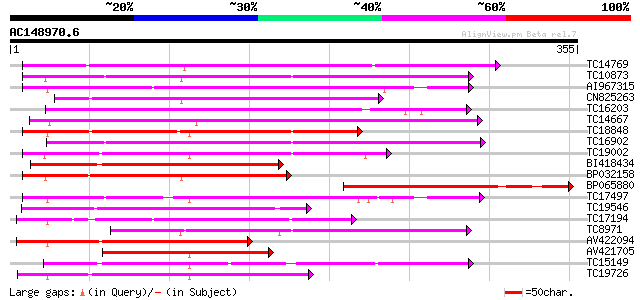
Score E
Sequences producing significant alignments: (bits) Value
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 184 3e-47
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 179 8e-46
AI967315 168 1e-42
CN825263 161 2e-40
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 161 2e-40
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 160 2e-40
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 160 2e-40
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 157 2e-39
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 156 6e-39
BI418434 151 2e-37
BP032158 151 2e-37
BP065880 149 7e-37
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 145 8e-36
TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine k... 141 2e-34
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 140 2e-34
TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P... 137 4e-33
AV422094 134 2e-32
AV421705 132 9e-32
TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like pro... 128 1e-30
TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase... 127 2e-30
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 184 bits (466), Expect = 3e-47
Identities = 115/303 (37%), Positives = 164/303 (53%), Gaps = 4/303 (1%)
Frame = +3
Query: 9 FTSQQLRIATDNYSNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVGTI 68
FT + + +AT+ Y L+G GGFG+VY+G N+G VAVKV R S+ + +F E+ +
Sbjct: 2133 FTLEYIEVATERYKTLIGEGGFGSVYRGTLNDGQEVAVKV-RSSTSTQGTREFDNELNLL 2309
Query: 69 GRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHE---KNALGYEKLHEIAIGTA 125
I H NLV L G+C E + LVY +M NGSL L E + L + IA+G A
Sbjct: 2310 SAIQHENLVPLLGYCNESDQQILVYPFMSNGSLQDRLYGEPAKRKILDWPTRLSIALGAA 2489
Query: 126 RGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTPG 185
RG+AYLH ++H DIK NILLD + KVADFG +K +E RGT G
Sbjct: 2490 RGLAYLHTFPGRSVIHRDIKSSNILLDHSMCAKVADFGFSKYAPQEGDSYVSLEVRGTAG 2669
Query: 186 YAAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQEWFPIWVWKKFDAGLLE 245
Y PE + ++ K DV+SFG++L EIV R L+IK + EW + + G
Sbjct: 2670 YLDPEYYKTQQLSEKSDVFSFGVVLLEIVSGREPLNIKRPRT-EWSLVEWATPYIRGSKV 2846
Query: 246 EAMIVCGIE-EKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLEIPKTFNPFQH 304
+ ++ GI+ + E R+V+VAL C++ RP M +V+ LE +L I + +
Sbjct: 2847 DEIVDPGIKGGYHAEAMWRVVEVALQCLEPFSTYRPSMVAIVRELEDALIIENNASEYMK 3026
Query: 305 LID 307
ID
Sbjct: 3027 SID 3035
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 179 bits (453), Expect = 8e-46
Identities = 108/289 (37%), Positives = 163/289 (56%), Gaps = 7/289 (2%)
Frame = +1
Query: 9 FTSQQLRIATDNY--SNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVG 66
F ++L AT N+ +NL+G GGFG VYKG G VAVK L + +E F+ EV
Sbjct: 415 FGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQE-FVMEVL 591
Query: 67 TIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLL---HEKNALGYEKLHEIAIG 123
+ +HH NLV+L G+C + + LVYEYM GSL+ +L H+K L + ++A+G
Sbjct: 592 MLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVG 771
Query: 124 TARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCN-RENTHITMTGGRG 182
ARG+ YLH + +++ D+K NILLD FNPK++DFGLAKL +NTH++ T G
Sbjct: 772 AARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVS-TRVMG 948
Query: 183 TPGYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQEWFPIWVWKKFDAG 242
T GY APE M +T K D+YSFG++L E++ RR +D ++ W F
Sbjct: 949 TYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDR 1128
Query: 243 LLEEAMIVCGIEEK-NREIAERMVKVALWCVQYRQQLRPMMSDVVKMLE 290
M+ ++ + + + + C+Q + + RP+++D+V LE
Sbjct: 1129RRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALE 1275
>AI967315
Length = 1308
Score = 168 bits (425), Expect = 1e-42
Identities = 105/290 (36%), Positives = 164/290 (56%), Gaps = 8/290 (2%)
Frame = +1
Query: 9 FTSQQLRIATDNYS--NLLGSGGFGTVYKGIFNNGTMVAVKVL-RGSSDKKIEEQFMAEV 65
F+ ++L AT+ +S N++G GG+ VYKG +G +AVK L R D++ E++F+ E+
Sbjct: 112 FSYEELFHATNGFSSENMVGKGGYAEVYKGRLESGDEIAVKRLTRTCRDERKEKEFLTEI 291
Query: 66 GTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA-LGYEKLHEIAIGT 124
GTIG + H N++ L G C + L LV+E GS+ + EK A L ++ ++I +GT
Sbjct: 292 GTIGHVCHSNVMPLLGCCIDNGLY-LVFELSTVGSVASLIHDEKMAPLDWKTRYKIVLGT 468
Query: 125 ARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTP 184
ARG+ YLH+ C+ RI+H DIK NILL +F P+++DFGLAK + TH ++ GT
Sbjct: 469 ARGLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPSQWTHHSIAPIEGTF 648
Query: 185 GYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQEWF-PI---WVWKKFD 240
G+ APE +M + K DV++FG+ L E++ R+ +D + W PI W +K
Sbjct: 649 GHLAPEYYMHGVVDEKTDVFAFGVFLLEVISGRKPVDGSHQSLHTWAKPILSKWEIEKLV 828
Query: 241 AGLLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLE 290
LE V R+ A C++ RP MS+V++++E
Sbjct: 829 DPRLEGCYDVTQF--------NRVAFAASLCIRASSTWRPTMSEVLEVME 954
>CN825263
Length = 663
Score = 161 bits (407), Expect = 2e-40
Identities = 90/209 (43%), Positives = 123/209 (58%), Gaps = 3/209 (1%)
Frame = +1
Query: 29 GFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVGTIGRIHHFNLVKLFGFCFEKNL 88
GFG VYKGI N+G VAVK+L+ D++ +F+AEV + R+HH NLVKL G C EK
Sbjct: 1 GFGLVYKGILNDGRDVAVKILK-RDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQT 177
Query: 89 IALVYEYMGNGSLDRYLL---HEKNALGYEKLHEIAIGTARGIAYLHELCEHRIVHYDIK 145
L+YE + NGS++ +L E L + +IA+G ARG+AYLHE ++H D K
Sbjct: 178 RCLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRDFK 357
Query: 146 PGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTPGYAAPELWMPFPITHKCDVYS 205
NILL+ +F PKV+DFGLA+ E T GT GY APE M + K DVYS
Sbjct: 358 SSNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSDVYS 537
Query: 206 FGMLLFEIVGRRRNLDIKNTESQEWFPIW 234
+G++L E++ + +D+ QE W
Sbjct: 538 YGVVLLELLTGTKPVDLSQPPGQENLVTW 624
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 161 bits (407), Expect = 2e-40
Identities = 99/276 (35%), Positives = 146/276 (52%), Gaps = 9/276 (3%)
Frame = +2
Query: 23 NLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVGTIGRIHHFNLVKLFGF 82
N++G GG G VY+G NGT VA+K L G + + F AE+ T+G+I H N+++L G+
Sbjct: 2204 NIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRLLGY 2383
Query: 83 CFEKNLIALVYEYMGNGSLDRYLLHEKNA-LGYEKLHEIAIGTARGIAYLHELCEHRIVH 141
K+ L+YEYM NGSL +L K L +E ++IA+ ARG+ Y+H C I+H
Sbjct: 2384 VSNKDTNLLLYEYMPNGSLGEWLHGAKGGHLRWEMRYKIAVEAARGLCYMHHDCSPLIIH 2563
Query: 142 YDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTPGYAAPELWMPFPITHKC 201
D+K NILLD +F VADFGLAK +M+ G+ GY APE + K
Sbjct: 2564 RDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKS 2743
Query: 202 DVYSFGMLLFEIVGRRRNLDIKNTESQEWFPIWVWKKFDAGLLEE----AMIVCGIEEK- 256
DVYSFG++L E++ R+ + E + I W L + A+++ ++ +
Sbjct: 2744 DVYSFGVVLLELIIGRKPVG----EFGDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRL 2911
Query: 257 ---NREIAERMVKVALWCVQYRQQLRPMMSDVVKML 289
M +A+ CV+ RP M +VV ML
Sbjct: 2912 SGYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHML 3019
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 160 bits (406), Expect = 2e-40
Identities = 99/294 (33%), Positives = 150/294 (50%), Gaps = 10/294 (3%)
Frame = +2
Query: 13 QLRIATDNYSN--LLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVGTIGR 70
+L+ TDN+ + L+G G +G VY N+G VAVK L SS+ + +F+ +V + R
Sbjct: 335 ELKEKTDNFGSKALIGEGSYGRVYYATLNDGNAVAVKKLDVSSEPETNNEFLTQVSMVSR 514
Query: 71 IHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNALGYE--------KLHEIAI 122
+ + N V+L G+C E NL L YE+ GSL L K G + + IA+
Sbjct: 515 LKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLDWIQRVRIAV 694
Query: 123 GTARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRG 182
ARG+ YLHE + I+H DI+ N+L+ ++ K+ADF L+ + T G
Sbjct: 695 DAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDMAARLHSTRVLG 874
Query: 183 TPGYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQEWFPIWVWKKFDAG 242
T GY APE M +T K DVYSFG++L E++ R+ +D Q+ W +
Sbjct: 875 TFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQSLVTWATPRLSED 1054
Query: 243 LLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLEIP 296
+++ + E + ++ VA CVQY + RP MS VVK L+ L+ P
Sbjct: 1055KVKQCVDPKLKGEYPPKGVAKLAAVAALCVQYEAEFRPNMSIVVKALQPLLKTP 1216
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 160 bits (406), Expect = 2e-40
Identities = 97/219 (44%), Positives = 135/219 (61%), Gaps = 6/219 (2%)
Frame = +2
Query: 9 FTSQQLRIATDNYS--NLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVG 66
FT ++L AT +S N LG GGFG+VY G ++G +AVK L+ + + K E +F EV
Sbjct: 266 FTYKELHAATGGFSDDNKLGEGGFGSVYWGRTSDGLQIAVKKLK-AMNSKAEMEFAVEVE 442
Query: 67 TIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA----LGYEKLHEIAI 122
+GR+ H NL+ L G+C + +VY+YM N SL +L H + A L ++K +IAI
Sbjct: 443 VLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHL-HGQFAVEVQLNWQKRMKIAI 619
Query: 123 GTARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRG 182
G+A GI YLH I+H DIK N+LL+ +F P VADFG AKL +H+T T +G
Sbjct: 620 GSAEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPLVADFGFAKLIPEGVSHMT-TRVKG 796
Query: 183 TPGYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLD 221
T GY APE M ++ CDVYSFG+LL E+V R+ ++
Sbjct: 797 TLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIE 913
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 157 bits (398), Expect = 2e-39
Identities = 100/277 (36%), Positives = 158/277 (56%), Gaps = 2/277 (0%)
Frame = +3
Query: 24 LLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVGTIGRIHHFNLVKLFGFC 83
LLG GGFG VY G + GT VA+K S++ + E F E+ + ++ H +LV L G+C
Sbjct: 12 LLGVGGFGKVYYGEVDGGTKVAIKRGNPLSEQGVHE-FQTEIEMLSKLRHRHLVSLIGYC 188
Query: 84 FEKNLIALVYEYMGNGSLDRYLLH-EKNALGYEKLHEIAIGTARGIAYLHELCEHRIVHY 142
E + LVY++M G+L +L +K L +++ EI IG ARG+ YLH ++ I+H
Sbjct: 189 EENTEMILVYDHMAYGTLREHLYKTQKPPLPWKQRLEICIGAARGLHYLHTGAKYTIIHR 368
Query: 143 DIKPGNILLDGNFNPKVADFGLAKL-CNRENTHITMTGGRGTPGYAAPELWMPFPITHKC 201
D+K NILLD + KV+DFGL+K +NTH++ T +G+ GY PE + +T K
Sbjct: 369 DVKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVS-TVVKGSFGYLDPEYFRRQQLTDKS 545
Query: 202 DVYSFGMLLFEIVGRRRNLDIKNTESQEWFPIWVWKKFDAGLLEEAMIVCGIEEKNREIA 261
DVYSFG++LFEI+ R L+ + Q W ++ G+L++ + + E
Sbjct: 546 DVYSFGVVLFEILCARPALNPSLAKEQVSLAEWASHCYNKGILDQILDPYLKGKIAPECF 725
Query: 262 ERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLEIPKT 298
++ + A+ CV + RP M DV+ LE +L++ ++
Sbjct: 726 KKFAETAMKCVSDQGIERPSMGDVLWNLEFALQLQES 836
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 156 bits (394), Expect = 6e-39
Identities = 98/241 (40%), Positives = 139/241 (57%), Gaps = 10/241 (4%)
Frame = +1
Query: 9 FTSQQLRIATDN--YSNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVG 66
F+ ++L AT+N Y N LG GGFG+VY G +G+ +AVK L+ S+K + +F EV
Sbjct: 28 FSLKELHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKVWSNKA-DMEFAVEVE 204
Query: 67 TIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA---LGYEKLHEIAIG 123
+ R+ H NL+ L G+C E +VY+YM N SL +L + ++ L + + IAIG
Sbjct: 205 ILARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECLLDWNRRMNIAIG 384
Query: 124 TARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGT 183
+A GI YLH I+H DIK N+LLD +F +VADFG AKL TH+T T +GT
Sbjct: 385 SAEGIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPDGATHVT-TRVKGT 561
Query: 184 PGYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLD-----IKNTESQEWFPIWVWKK 238
GY APE M CDV+SFG+LL E+ ++ L+ +K + + P+ KK
Sbjct: 562 LGYLAPEYAMLGKANECCDVFSFGILLLELASGKKPLEKLSSTVKRSINDWALPLACAKK 741
Query: 239 F 239
F
Sbjct: 742 F 744
>BI418434
Length = 478
Score = 151 bits (381), Expect = 2e-37
Identities = 81/158 (51%), Positives = 106/158 (66%)
Frame = +2
Query: 14 LRIATDNYSNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVGTIGRIHH 73
L++AT +S +G GGFGTV++G ++ ++VAVK L E++F AEV TIG I H
Sbjct: 2 LQLATRGFSEKVGHGGFGTVFQGELSDSSLVAVKRLERPGGG--EKEFRAEVSTIGNIQH 175
Query: 74 FNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNALGYEKLHEIAIGTARGIAYLHE 133
NLV+L GFC E + LVYEYM NG+L YL + L ++ ++AIGTA+GIAYLHE
Sbjct: 176 VNLVRLRGFCSENSHRLLVYEYMHNGALSAYLRKDGPCLSWDVRFQVAIGTAKGIAYLHE 355
Query: 134 LCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRE 171
I+H DIKP NILLD +F KV+DFGLAKL R+
Sbjct: 356 ELRCCILHCDIKPENILLDSDFTAKVSDFGLAKLIGRD 469
>BP032158
Length = 555
Score = 151 bits (381), Expect = 2e-37
Identities = 81/173 (46%), Positives = 112/173 (63%), Gaps = 5/173 (2%)
Frame = +2
Query: 9 FTSQQLRIATDNY--SNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVG 66
F+ +Q++ AT+N+ +N +G GGFG VYKG+ + G ++AVK L S K+ +F+ E+G
Sbjct: 14 FSLRQIKAATNNFDPANKIGEGGFGPVYKGVLSEGDVIAVKQL-SSKSKQGNREFINEIG 190
Query: 67 TIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLL---HEKNALGYEKLHEIAIG 123
I + H NLVKL+G C E N + LVYEYM N SL R L +K L + +I +G
Sbjct: 191 MISALQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGNEEQKLNLNWRTRMKICVG 370
Query: 124 TARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHIT 176
A+G+AYLHE +IVH DIK N+LLD + K++DFGLAKL ENTHI+
Sbjct: 371 IAKGLAYLHEESRLKIVHRDIKATNVLLDKDLKAKISDFGLAKLDEEENTHIS 529
>BP065880
Length = 509
Score = 149 bits (376), Expect = 7e-37
Identities = 80/144 (55%), Positives = 96/144 (66%)
Frame = -1
Query: 210 LFEIVGRRRNLDIKNTESQEWFPIWVWKKFDAGLLEEAMIVCGIEEKNREIAERMVKVAL 269
LFEI+ RRRN D ESQEWFP W W+KFD+G LEE I C EEKN EI ERM KVAL
Sbjct: 509 LFEIIVRRRNFDTSLPESQEWFPRWXWEKFDSGKLEELRIACRNEEKNMEIVERMSKVAL 330
Query: 270 WCVQYRQQLRPMMSDVVKMLEGSLEIPKTFNPFQHLIDETKFTTHSDQESNTYTTSVSSV 329
WCVQYR +LRP+MS VV+MLEG +EIPK NPFQ ++ T + ++ T+VS
Sbjct: 329 WCVQYRSELRPLMSGVVRMLEGVVEIPKPLNPFQTMM----VGTLPNPTNHPSDTTVS-- 168
Query: 330 MVSDSNIVCATPIMRKYEIELASS 353
S CATPI+ ++ELASS
Sbjct: 167 ----SGCGCATPILTILDVELASS 108
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 145 bits (367), Expect = 8e-36
Identities = 108/302 (35%), Positives = 156/302 (50%), Gaps = 13/302 (4%)
Frame = +1
Query: 9 FTSQQLRIATDNYS--NLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVG 66
F + AT+++S N LG GGFG VYKG+ NG +AVK L +S + +EE F E+
Sbjct: 1636 FDFSTISSATNHFSLSNKLGEGGFGPVYKGLLANGQEIAVKRLSNTSGQGMEE-FKNEIK 1812
Query: 67 TIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA--LGYEKLHEIAIGT 124
I R+ H NLVKLFG ++ + + M + LL + + + K +I G
Sbjct: 1813 LIARLQHRNLVKLFGCSVHQDENSHANKKM------KILLDSTRSKLVDWNKRLQIIDGI 1974
Query: 125 ARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTP 184
ARG+ YLH+ RI+H D+K NILLD NPK++DFGLA++ + GT
Sbjct: 1975 ARGLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIFIGDQVEARTKRVMGTY 2154
Query: 185 GYAAPELWMPFPITHKCDVYSFGMLLFEIVGRR---RNLDIK---NTESQEWFPIWVWKK 238
GY PE + + K DV+SFG+++ EI+ + R D N S W +W+ ++
Sbjct: 2155 GYMPPEYAVHGSFSIKSDVFSFGVIVLEIISGKKIGRFYDPHHHLNLLSHAW-RLWIEER 2331
Query: 239 ---FDAGLLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLEI 295
LL++ +I I R + VAL CVQ R + RP M +V ML G E+
Sbjct: 2332 PLELVDELLDDPVIPTEI--------LRYIHVALLCVQRRPENRPDMLSIVLMLNGEKEL 2487
Query: 296 PK 297
PK
Sbjct: 2488 PK 2493
>TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine kinase ,
partial (46%)
Length = 731
Score = 141 bits (355), Expect = 2e-34
Identities = 78/182 (42%), Positives = 110/182 (59%)
Frame = +1
Query: 8 RFTSQQLRIATDNYSNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVGT 67
+++ + ++ AT N++ +LG G FGTVYK G +VAVKVL +S K+ E +F EV
Sbjct: 196 KYSYKDIQKATQNFTTILGQGSFGTVYKATMPTGEVVAVKVLAPNS-KQGEHEFQTEVHL 372
Query: 68 IGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNALGYEKLHEIAIGTARG 127
+GR+HH NLV L GFC +K LVY++M NGSL L E+ L +++ +IA+ + G
Sbjct: 373 LGRLHHRNLVNLVGFCVDKGQRILVYQFMSNGSLANLLYGEEKELSWDERLQIAMDISHG 552
Query: 128 IAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTPGYA 187
I YLHE ++H D+K NILLD + VADFGL+K E +G +GT GY
Sbjct: 553 IEYLHEGAVPPVIHRDLKSANILLDDSMRAMVADFGLSK---EEIFDGRNSGLKGTYGYM 723
Query: 188 AP 189
P
Sbjct: 724 DP 729
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 140 bits (354), Expect = 2e-34
Identities = 86/216 (39%), Positives = 124/216 (56%), Gaps = 3/216 (1%)
Frame = +1
Query: 5 KPIRFTSQQLRIATDNYS--NLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFM 62
K + F+ Q+L AT+N+S N +G GGFG VY G A+K + D + +F+
Sbjct: 1039 KSMEFSYQELAKATNNFSLDNKIGQGGFGAVYYAELR-GKKTAIKKM----DVQASTEFL 1203
Query: 63 AEVGTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHE-KNALGYEKLHEIA 121
E+ + +HH NLV+L G+C E +L LVYE++ NG+L +YL K L + +IA
Sbjct: 1204 CELKVLTHVHHLNLVRLIGYCVEGSLF-LVYEHIDNGNLGQYLHGSGKEPLPWSSRVQIA 1380
Query: 122 IGTARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGR 181
+ ARG+ Y+HE +H D+K NIL+D N KVADFGL KL N+ + T
Sbjct: 1381 LDAARGLEYIHEHTVPVYIHRDVKSANILIDKNLRGKVADFGLTKLIEVGNSTL-QTRLV 1557
Query: 182 GTPGYAAPELWMPFPITHKCDVYSFGMLLFEIVGRR 217
GT GY PE I+ K DVY+FG++LFE++ +
Sbjct: 1558 GTFGYMPPEYAQYGDISPKIDVYAFGVVLFELISAK 1665
>TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P14.15
{Arabidopsis thaliana;}, partial (37%)
Length = 1079
Score = 137 bits (344), Expect = 4e-33
Identities = 88/230 (38%), Positives = 126/230 (54%), Gaps = 4/230 (1%)
Frame = +3
Query: 64 EVGTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYL--LHEKNALGYEKLHEIA 121
EV + +I H NLVKL GF + N L+ EY+ NG+L +L L K L + + EIA
Sbjct: 6 EVELLAKIDHRNLVKLLGFIDKGNERILITEYVPNGTLREHLDGLRGK-ILDFNQRLEIA 182
Query: 122 IGTARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKL--CNRENTHITMTG 179
I A G+ YLH E +I+H D+K NILL + KVADFG A+L N + THI+ T
Sbjct: 183 IDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFARLGPVNGDQTHIS-TK 359
Query: 180 GRGTPGYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQEWFPIWVWKKF 239
+GT GY PE +T K DVYSFG+LL EI+ RR +++K + W ++K+
Sbjct: 360 VKGTVGYLDPEYMKTHQLTPKSDVYSFGILLLEILTGRRPVELKKAADERVTLRWAFRKY 539
Query: 240 DAGLLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKML 289
+ G + E M E N ++ +M+ ++ C + RP M V + L
Sbjct: 540 NEGSVVELMDPLMEEAVNADVLMKMLDLSFQCAAPIRTDRPNMKSVGEQL 689
>AV422094
Length = 462
Score = 134 bits (338), Expect = 2e-32
Identities = 72/150 (48%), Positives = 99/150 (66%), Gaps = 2/150 (1%)
Frame = +2
Query: 5 KPIRFTSQQLRIATDNYS--NLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFM 62
KP F+ +L+ AT++++ N LG GGFG VYKGI N+GT++AVK L S + + QF+
Sbjct: 2 KPYTFSYSELKNATNDFNIDNKLGEGGFGPVYKGILNDGTVIAVKQLSLGSHQG-KSQFI 178
Query: 63 AEVGTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNALGYEKLHEIAI 122
AE+ TI + H NLVKL+G C E + LVYEY+ N SLD+ L + +L + ++I +
Sbjct: 179 AEIATISAVQHRNLVKLYGCCIEGSKRLLVYEYLENKSLDQALFGKALSLNWSTRYDICL 358
Query: 123 GTARGIAYLHELCEHRIVHYDIKPGNILLD 152
G ARG+ YLHE RIVH D+K NILLD
Sbjct: 359 GVARGLTYLHEESRLRIVHRDVKASNILLD 448
>AV421705
Length = 341
Score = 132 bits (332), Expect = 9e-32
Identities = 60/109 (55%), Positives = 87/109 (79%), Gaps = 2/109 (1%)
Frame = +3
Query: 59 EQFMAEVGTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA--LGYEK 116
+ F++EV TIGRIHH N+V+L G+C + ALVYE+M NGSLD+Y+ ++ + L Y++
Sbjct: 15 QDFISEVATIGRIHHINVVRLVGYCVDGKKRALVYEFMPNGSLDKYIFSKEGSAWLSYDQ 194
Query: 117 LHEIAIGTARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLA 165
++EI++G ARG+AYLH+ C+ +I+H+DIKP NILLD +F PKV+DFGLA
Sbjct: 195 IYEISLGIARGVAYLHQGCDMQILHFDIKPHNILLDKDFIPKVSDFGLA 341
>TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like protein kinase
protein (AT3g17840/MEB5_6), partial (45%)
Length = 1489
Score = 128 bits (322), Expect = 1e-30
Identities = 93/275 (33%), Positives = 142/275 (50%), Gaps = 6/275 (2%)
Frame = +3
Query: 22 SNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVGTIGRIHHFNLVKLFG 81
+ +LG G FGT YK + G +VAVK L+ + E++F ++ T+G + H +LV L
Sbjct: 285 AEVLGKGTFGTAYKAVLETGLVVAVKRLKDVTIS--EKEFKDKIETVGAMDHQSLVPLRA 458
Query: 82 FCFEKNLIALVYEYMGNGSLDRYLLHEKNA----LGYEKLHEIAIGTARGIAYLHELCEH 137
+ F ++ LVY+YM GSL L K A L +E IA+G ARGI YLH +
Sbjct: 459 YYFSRDEKLLVYDYMPMGSLSALLHGNKGAGRTPLNWEIRSGIALGAARGIEYLHSQGPN 638
Query: 138 RIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTPGYAAPELWMPFPI 197
+ H +IK NILL ++ KV+DFGLA L +T GY APE+ P +
Sbjct: 639 -VSHGNIKASNILLTKSYEAKVSDFGLAHLVGPSST------PNRVAGYRAPEVTDPRKV 797
Query: 198 THKCDVYSFGMLLFEIV-GRRRNLDIKNTESQEWFPIWVWKKFDAGLLEEAMIVCGIEEK 256
+ K DVYSFG+LL E++ G+ + N E + P WV E + + +
Sbjct: 798 SQKADVYSFGVLLLELLTGKAPTHALLNEEGVD-LPRWVQSLVREEWTSEVFDLELLRYQ 974
Query: 257 N-REIAERMVKVALWCVQYRQQLRPMMSDVVKMLE 290
N E +++++A+ C RP +++V + +E
Sbjct: 975 NVEEEMVQLLQLAVDCAAPYPDRRPSIAEVTRSIE 1079
>TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase, partial
(30%)
Length = 728
Score = 127 bits (320), Expect = 2e-30
Identities = 76/195 (38%), Positives = 116/195 (58%), Gaps = 10/195 (5%)
Frame = +3
Query: 6 PIRFTSQQLRIATDNYS--NLLGSGGFGTVYKGIFNNGTM-VAVKVLRGSSDK-KIEEQF 61
P F +L+ AT+N+ + LG GG+G VY+G+ + VAVK+ S DK K + F
Sbjct: 123 PREFNYVELKKATNNFDEKHKLGQGGYGVVYRGMLPKEKLEVAVKMF--SRDKMKSTDDF 296
Query: 62 MAEVGTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA-----LGYEK 116
++E+ I R+ H +LV+L G+C + ++ LVY+YM NGSLD ++ E+ L +
Sbjct: 297 LSELIIINRLRHKHLVRLQGWCHKNGVLLLVYDYMPNGSLDSHIFCEEGGTITTPLSWPL 476
Query: 117 LHEIAIGTARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAK-LCNRENTHI 175
++I G A + YLH + ++VH D+K NI+LD FN K+ DFGLA+ L N + ++
Sbjct: 477 RYKIISGVASALNYLHNEYDQKVVHRDLKASNIMLDSEFNAKLGDFGLARALENEKISYT 656
Query: 176 TMTGGRGTPGYAAPE 190
+ G GT GY APE
Sbjct: 657 ELEGVHGTMGYIAPE 701
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.138 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,972,602
Number of Sequences: 28460
Number of extensions: 84504
Number of successful extensions: 842
Number of sequences better than 10.0: 293
Number of HSP's better than 10.0 without gapping: 669
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 672
length of query: 355
length of database: 4,897,600
effective HSP length: 91
effective length of query: 264
effective length of database: 2,307,740
effective search space: 609243360
effective search space used: 609243360
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC148970.6


