

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
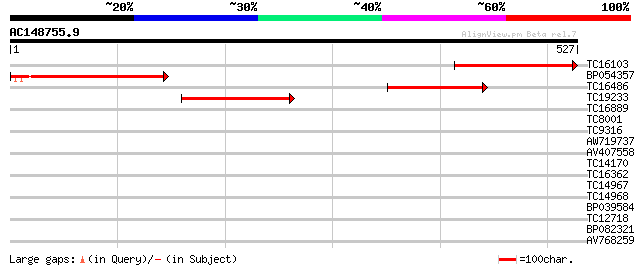
Query= AC148755.9 + phase: 0
(527 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC16103 similar to UP|Q9FEK8 (Q9FEK8) Ferrochelatase (Protoheme... 206 1e-53
BP054357 179 1e-45
TC16486 similar to UP|HMZ1_ARATH (P42043) Ferrochelatase I, chlo... 159 8e-40
TC19233 similar to UP|HEMZ_CUCSA (P42044) Ferrochelatase II, chl... 154 4e-38
TC16889 weakly similar to UP|PSBS_ARATH (Q9XF91) Photosystem II ... 35 0.030
TC8001 similar to UP|Q8LC02 (Q8LC02) Lil3 protein, partial (56%) 32 0.26
TC9316 similar to UP|Q94F86 (Q94F86) Early light inducible prote... 32 0.33
AW719737 32 0.33
AV407558 31 0.57
TC14170 similar to UP|PSBS_LYCES (P54773) Photosystem II 22 kDa ... 30 0.97
TC16362 similar to UP|HCD2_HUMAN (Q99714) 3-hydroxyacyl-CoA dehy... 30 1.3
TC14967 28 2.8
TC14968 similar to UP|Q8TTE6 (Q8TTE6) Bacterial extracellular so... 28 3.7
BP039584 28 4.8
TC12718 27 8.2
BP082321 27 8.2
AV768259 27 8.2
>TC16103 similar to UP|Q9FEK8 (Q9FEK8) Ferrochelatase (Protoheme
ferro-lyase) (Heme synthetase) , partial (21%)
Length = 666
Score = 206 bits (523), Expect = 1e-53
Identities = 97/114 (85%), Positives = 104/114 (91%)
Frame = +3
Query: 414 WGRVPALGCEPTFISDLADAVIESLPYVGAMAVSNLEARQSLVPLGSVEELLAAYDSQRR 473
WGRVPALGCEP+FISDLADAVIESLPYVGAMAVS+LEARQSLVPLGSVEELLA YDS+RR
Sbjct: 6 WGRVPALGCEPSFISDLADAVIESLPYVGAMAVSDLEARQSLVPLGSVEELLATYDSRRR 185
Query: 474 ELPPPILVWEWGWTKSAETWNGRAAMIAVLLLLFLEVTTGEGFLHQWGILPLFR 527
ELPPP +VWEWGWTKS+ETWNGR AM+A LLL+F EVTT FLHQWGILP R
Sbjct: 186 ELPPPFVVWEWGWTKSSETWNGRVAMLAFLLLMFFEVTTEGSFLHQWGILPSLR 347
>BP054357
Length = 566
Score = 179 bits (453), Expect = 1e-45
Identities = 99/154 (64%), Positives = 112/154 (72%), Gaps = 7/154 (4%)
Frame = +3
Query: 1 MNSP--IRAPS-----TSSCSYIRPHSCRTCASRNFKLPLLLPQAICTAQKLHRCSGGHT 53
MNSP I APS +SSCS+ P C T ASRNFK PL LPQ + T+QK++ CSG
Sbjct: 90 MNSPASIHAPSLPSSSSSSCSHRSP-PCLTHASRNFKFPLSLPQPMFTSQKIYCCSGADM 266
Query: 54 EASSNVNPLKTCIVGKFSPGWSEAQPLVSKQSLNRHLLPVEALVTSTTQDVSDTPLIGDD 113
EAS N NP+K I + SP W EAQPL+S QSLN++L V AL T T QDVSDT LIGDD
Sbjct: 267 EASVNTNPMKLHIETRCSPRWFEAQPLISNQSLNKNLFSVRALATPTAQDVSDTTLIGDD 446
Query: 114 KIGVLLLNLGGPETLDDVQPFLFNLFADPDIIRL 147
K GVLLLNLGGPETL+DVQPFLFNLFADP I+ L
Sbjct: 447 KTGVLLLNLGGPETLEDVQPFLFNLFADPVIVIL 548
>TC16486 similar to UP|HMZ1_ARATH (P42043) Ferrochelatase I,
chloroplast/mitochondrial precursor (Protoheme
ferro-lyase) (Heme synthetase) , partial (25%)
Length = 658
Score = 159 bits (403), Expect = 8e-40
Identities = 76/93 (81%), Positives = 86/93 (91%)
Frame = +3
Query: 352 QSRVGPVEWLKPYTDETIIELGKKGVKSLLAVPISFVSEHIETLEEIDVEYKELALESGI 411
QSRVGPV+WLKPYTDE ++ELGKKGVKSLLAVP+SFVSEHIETLEEID+EYKELALESG+
Sbjct: 3 QSRVGPVQWLKPYTDEVLVELGKKGVKSLLAVPVSFVSEHIETLEEIDMEYKELALESGV 182
Query: 412 ENWGRVPALGCEPTFISDLADAVIESLPYVGAM 444
+NW RVPALG P+FI+DLADAVIE+LP AM
Sbjct: 183 KNWARVPALGLTPSFITDLADAVIEALPSATAM 281
>TC19233 similar to UP|HEMZ_CUCSA (P42044) Ferrochelatase II, chloroplast
precursor (Protoheme ferro-lyase) (Heme synthetase) ,
partial (22%)
Length = 337
Score = 154 bits (388), Expect = 4e-38
Identities = 72/105 (68%), Positives = 92/105 (87%)
Frame = +3
Query: 160 QFVSVLRAPKSKEGYASIGGGSPLRRMTDAQAEELRKSLFEKNVPANVYVGMRYWHPFTE 219
+ +S LRAPKSKE YASIGGGSPLR++TD QA ++K+L K + ++VYVGMRYW+PFTE
Sbjct: 3 KLISTLRAPKSKEAYASIGGGSPLRKITDDQALAIKKALEAKGLSSSVYVGMRYWYPFTE 182
Query: 220 EAIELIKRDGITKLVVLPLYPQFSISTSGSSLRLLESIFREDEYL 264
+A++ IKRDGIT+LVVLPLYPQFSIST+GSS+R+L+ +FRED YL
Sbjct: 183 DAVQQIKRDGITRLVVLPLYPQFSISTTGSSIRVLQHMFREDAYL 317
>TC16889 weakly similar to UP|PSBS_ARATH (Q9XF91) Photosystem II 22 kDa
protein, chloroplast precursor (CP22), partial (14%)
Length = 415
Score = 35.0 bits (79), Expect = 0.030
Identities = 19/40 (47%), Positives = 24/40 (59%)
Frame = +2
Query: 484 WGWTKSAETWNGRAAMIAVLLLLFLEVTTGEGFLHQWGIL 523
+G+T E NG+AA I LLLL E+ TG+G L G L
Sbjct: 56 FGFTVKNEIINGKAAAIGFLLLLDFELLTGKGLLKGTGFL 175
>TC8001 similar to UP|Q8LC02 (Q8LC02) Lil3 protein, partial (56%)
Length = 1039
Score = 32.0 bits (71), Expect = 0.26
Identities = 18/46 (39%), Positives = 23/46 (49%), Gaps = 5/46 (10%)
Frame = +2
Query: 479 ILVWEWGWTK-----SAETWNGRAAMIAVLLLLFLEVTTGEGFLHQ 519
I+ W W W K AE NGRAAMI + ++ TG G + Q
Sbjct: 413 IVPW-WAWIKRFHLPEAERLNGRAAMIGFFMAYLVDSLTGVGLVDQ 547
>TC9316 similar to UP|Q94F86 (Q94F86) Early light inducible protein,
partial (84%)
Length = 831
Score = 31.6 bits (70), Expect = 0.33
Identities = 13/27 (48%), Positives = 16/27 (59%)
Frame = +1
Query: 487 TKSAETWNGRAAMIAVLLLLFLEVTTG 513
T AE WNGR AM+ ++ L F E G
Sbjct: 598 TSDAELWNGRLAMLGLIALAFTEYVKG 678
>AW719737
Length = 481
Score = 31.6 bits (70), Expect = 0.33
Identities = 22/70 (31%), Positives = 32/70 (45%), Gaps = 13/70 (18%)
Frame = +3
Query: 179 GGSPLRRMTDAQAEELRKSLFEK-------------NVPANVYVGMRYWHPFTEEAIELI 225
G P + + D + E LRK EK VPA ++ ++ P + I+LI
Sbjct: 234 GKLPAKSVNDEKLEVLRKDEKEKIKKLFSKYISFCGKVPAEIFEVEKFDEPIQKRTIDLI 413
Query: 226 KRDGITKLVV 235
GITKLV+
Sbjct: 414 FGLGITKLVI 443
>AV407558
Length = 310
Score = 30.8 bits (68), Expect = 0.57
Identities = 13/32 (40%), Positives = 19/32 (58%)
Frame = +3
Query: 491 ETWNGRAAMIAVLLLLFLEVTTGEGFLHQWGI 522
+ W GR AMI + +EV TG+G L +G+
Sbjct: 180 DVWLGRLAMIGFAAGITVEVATGKGLLENFGV 275
>TC14170 similar to UP|PSBS_LYCES (P54773) Photosystem II 22 kDa protein,
chloroplast precursor (CP22), partial (81%)
Length = 1201
Score = 30.0 bits (66), Expect = 0.97
Identities = 16/39 (41%), Positives = 22/39 (56%)
Frame = +3
Query: 484 WGWTKSAETWNGRAAMIAVLLLLFLEVTTGEGFLHQWGI 522
+G+TKS E + GR A + L E+ TG+G L Q I
Sbjct: 720 FGFTKSNELFVGRLAQLGFAFSLIGEIITGKGALAQLNI 836
Score = 29.6 bits (65), Expect = 1.3
Identities = 16/35 (45%), Positives = 19/35 (53%)
Frame = +3
Query: 485 GWTKSAETWNGRAAMIAVLLLLFLEVTTGEGFLHQ 519
G+TK E + GR AMI L E TG+G L Q
Sbjct: 408 GFTKQNELFVGRVAMIGFAASLLGEALTGKGILAQ 512
>TC16362 similar to UP|HCD2_HUMAN (Q99714) 3-hydroxyacyl-CoA dehydrogenase
type II (Type II HADH) (Endoplasmic
reticulum-associated amyloid beta-peptide binding
protein) (Short-chain type dehydrogenase/reductase
XH98G2) , partial (7%)
Length = 613
Score = 29.6 bits (65), Expect = 1.3
Identities = 18/49 (36%), Positives = 26/49 (52%)
Frame = +1
Query: 48 CSGGHTEASSNVNPLKTCIVGKFSPGWSEAQPLVSKQSLNRHLLPVEAL 96
CS GH EA + + I GK W +Q L +KQ + H+LP+ A+
Sbjct: 334 CSSGHNEA*PLLTEICEIICGKLHRSWPSSQHL-NKQCWS-HVLPLPAI 474
>TC14967
Length = 655
Score = 28.5 bits (62), Expect = 2.8
Identities = 16/55 (29%), Positives = 27/55 (49%), Gaps = 2/55 (3%)
Frame = -2
Query: 6 RAPSTSSCSYIRPHSCRTCASRNFKLPLLLPQAICTAQKLHRCSG--GHTEASSN 58
+ PS + S++ C+T S K+P LL A+ + +G GHT+ + N
Sbjct: 291 KTPSLRTASFLPTKMCQTTTSHTDKIPSLLTDRNKPAKDIR*STGLPGHTKQTPN 127
>TC14968 similar to UP|Q8TTE6 (Q8TTE6) Bacterial extracellular
solute-binding family 3 protein, partial (3%)
Length = 503
Score = 28.1 bits (61), Expect = 3.7
Identities = 17/60 (28%), Positives = 29/60 (48%), Gaps = 2/60 (3%)
Frame = -1
Query: 1 MNSPIRAPSTSSCSYIRPHSCRTCASRNFKLPLLLPQAICTAQKLHRCSG--GHTEASSN 58
M+S + PS + S++ +C+T AS K+P A+ +G GHT+ + N
Sbjct: 293 MHSGRKTPSLKTASFLPTQTCQTTASHTDKIPSFWANRNNPAKDRR*STGVPGHTKQTPN 114
>BP039584
Length = 545
Score = 27.7 bits (60), Expect = 4.8
Identities = 15/61 (24%), Positives = 27/61 (43%)
Frame = -3
Query: 176 SIGGGSPLRRMTDAQAEELRKSLFEKNVPANVYVGMRYWHPFTEEAIELIKRDGITKLVV 235
S+ G P+ M + + L + V +GM+ W + E++KR+ I K +
Sbjct: 456 SVIAGLPMATMPLFAEQFYNEKLLVDVLGVGVSIGMKKWRNWNVVGDEIVKRENIVKAIS 277
Query: 236 L 236
L
Sbjct: 276 L 274
>TC12718
Length = 806
Score = 26.9 bits (58), Expect = 8.2
Identities = 16/43 (37%), Positives = 23/43 (53%)
Frame = +2
Query: 32 PLLLPQAICTAQKLHRCSGGHTEASSNVNPLKTCIVGKFSPGW 74
PL +CT + R +GG E SS VNP +C++ SP +
Sbjct: 17 PLHRVHLLCTV--VLRPAGGLNETSSFVNPSVSCVLEFSSPSF 139
>BP082321
Length = 377
Score = 26.9 bits (58), Expect = 8.2
Identities = 9/22 (40%), Positives = 10/22 (44%)
Frame = -1
Query: 472 RRELPPPILVWEWGWTKSAETW 493
RR+LP L W W W W
Sbjct: 323 RRQLPSRGLFWAWNWRSGTLNW 258
>AV768259
Length = 490
Score = 26.9 bits (58), Expect = 8.2
Identities = 14/33 (42%), Positives = 17/33 (51%)
Frame = -2
Query: 121 NLGGPETLDDVQPFLFNLFADPDIIRLPRLFSF 153
NLGG + LD Q F N + + RLP L F
Sbjct: 141 NLGGGQVLDVAQSFRVNQYKSLCVDRLPYLLIF 43
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.137 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,317,453
Number of Sequences: 28460
Number of extensions: 133089
Number of successful extensions: 728
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 715
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 728
length of query: 527
length of database: 4,897,600
effective HSP length: 94
effective length of query: 433
effective length of database: 2,222,360
effective search space: 962281880
effective search space used: 962281880
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC148755.9


