

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148755.7 - phase: 0
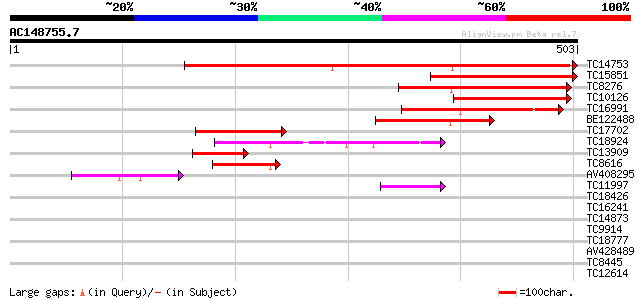
(503 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14753 similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, partial... 508 e-145
TC15851 226 5e-60
TC8276 similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, partial ... 189 9e-49
TC10126 174 4e-44
TC16991 similar to GB|AAN73306.1|25141223|BT002309 At3g51670/T18... 137 3e-33
BE122488 107 5e-24
TC17702 weakly similar to GB|AAN73306.1|25141223|BT002309 At3g51... 87 6e-18
TC18924 homologue to UP|Q94FN3 (Q94FN3) Phosphatidylinositol tra... 75 2e-14
TC13909 weakly similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, ... 72 3e-13
TC8616 UP|Q94FN1 (Q94FN1) Phosphatidylinositol transfer-like pro... 52 2e-07
AV408295 45 3e-05
TC11997 similar to UP|O48940 (O48940) Polyphosphoinositide bindi... 44 8e-05
TC18426 39 0.002
TC16241 weakly similar to UP|Q91TH4 (Q91TH4) T122, partial (7%) 36 0.013
TC14873 weakly similar to GB|BAB08982.1|9758453|AB008271 seleniu... 34 0.049
TC9914 weakly similar to GB|AAN73306.1|25141223|BT002309 At3g516... 33 0.083
TC18777 weakly similar to UP|Q9LDJ7 (Q9LDJ7) ESTs AU081555, part... 33 0.14
AV428489 32 0.24
TC8445 similar to UP|O80910 (O80910) At2g38410 protein, partial ... 32 0.32
TC12614 31 0.41
>TC14753 similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, partial (57%)
Length = 1472
Score = 508 bits (1309), Expect = e-145
Identities = 250/352 (71%), Positives = 293/352 (83%), Gaps = 4/352 (1%)
Frame = +1
Query: 156 ASLPLPPEQVSIYGIPLLADETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEEL 215
+ +P PE+VSI+GIPLLADE SDV+LLKFLRARDFKVK+AFTMIKNT+ WRKEFGI+ L
Sbjct: 13 SEIPSEPEEVSIWGIPLLADERSDVVLLKFLRARDFKVKDAFTMIKNTVRWRKEFGIDAL 192
Query: 216 MDEKLGDELEKVVYMHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFL 275
+DE LG E +KVV+ G DK GH VCYN++GEF++KELY KTFSD+EKR F+KWRIQ L
Sbjct: 193 VDEDLGSEWDKVVFTQGHDKLGHAVCYNVFGEFEDKELYQKTFSDDEKRQKFIKWRIQVL 372
Query: 276 EKSIRNLDFN--HGGVCTIVHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVF 333
EKS+R LDF+ + TIV VNDLK+SPG GKWELRQAT QALQL QDNYPEFVAKQVF
Sbjct: 373 EKSVRKLDFSPTPTAISTIVQVNDLKNSPGLGKWELRQATNQALQLLQDNYPEFVAKQVF 552
Query: 334 INVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKDG-- 391
INVPWWYLA +RMISPFLTQRTKSKFVFAGPSKS ETL YIAPE +PV+YGGLS++G
Sbjct: 553 INVPWWYLAFSRMISPFLTQRTKSKFVFAGPSKSAETLFKYIAPELVPVQYGGLSREGEQ 732
Query: 392 EFGNSDSVTEITIRPASKHTVEFPVTEKCLLSWEVRVIGWEVRYGAEFVPSNEGSYTVIV 451
EF +D VTE+TI+PA+KH VE PV+EK L+WE+RV+GW+V YGAEFVP E YTVIV
Sbjct: 733 EFTTADPVTEVTIKPATKHAVELPVSEKGHLAWELRVVGWDVNYGAEFVPGAEDGYTVIV 912
Query: 452 QKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKKLLYRLKTKTSLSD 503
QK RK+ ++E V+ NSFK+ E GKVVLTIDN +S+KKKLLYR KT T +SD
Sbjct: 913 QKNRKIGPADETVISNSFKVGEAGKVVLTIDNQTSKKKKLLYRSKT-TPISD 1065
>TC15851
Length = 684
Score = 226 bits (577), Expect = 5e-60
Identities = 112/130 (86%), Positives = 122/130 (93%)
Frame = +1
Query: 374 YIAPEQLPVKYGGLSKDGEFGNSDSVTEITIRPASKHTVEFPVTEKCLLSWEVRVIGWEV 433
YIAPEQLPVKYGGLSKDGEF NS++VTEITIRPA+KHTVEFPVTEKCLLSWE+RVIGW++
Sbjct: 4 YIAPEQLPVKYGGLSKDGEFENSEAVTEITIRPAAKHTVEFPVTEKCLLSWELRVIGWDI 183
Query: 434 RYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKKLLY 493
YGAEFVPS+EGSYTVIVQK RKVASSEE VL NSFKI EPGKVVLTI+NTSS+KKKLLY
Sbjct: 184 SYGAEFVPSSEGSYTVIVQKDRKVASSEEPVLFNSFKIEEPGKVVLTINNTSSKKKKLLY 363
Query: 494 RLKTKTSLSD 503
RLKTK + SD
Sbjct: 364 RLKTKLTPSD 393
>TC8276 similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, partial (42%)
Length = 1071
Score = 189 bits (480), Expect = 9e-49
Identities = 91/158 (57%), Positives = 119/158 (74%), Gaps = 5/158 (3%)
Frame = +3
Query: 346 MISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKD-----GEFGNSDSVT 400
MI+PFLTQRTKSKFVFAGPSKS +TL YI+PEQ+P++YGGLS D +F SD VT
Sbjct: 27 MINPFLTQRTKSKFVFAGPSKSPDTLFKYISPEQVPIQYGGLSVDFCDCNPDFSMSDPVT 206
Query: 401 EITIRPASKHTVEFPVTEKCLLSWEVRVIGWEVRYGAEFVPSNEGSYTVIVQKARKVASS 460
EI ++P +K TVE + EKC++ WE+RV+GWEV Y AEF P + YTVI+QKA K++ S
Sbjct: 207 EIPLKPNTKQTVEIAIYEKCIIVWELRVVGWEVSYSAEFKPDAKDGYTVIIQKATKMSPS 386
Query: 461 EEAVLCNSFKINEPGKVVLTIDNTSSRKKKLLYRLKTK 498
+E V+ N FK+ E GK++LT+DN + +KK+LLYR K
Sbjct: 387 DEPVVSNHFKVAELGKLLLTVDNPTLKKKRLLYRFNIK 500
>TC10126
Length = 588
Score = 174 bits (440), Expect = 4e-44
Identities = 85/105 (80%), Positives = 95/105 (89%)
Frame = +3
Query: 394 GNSDSVTEITIRPASKHTVEFPVTEKCLLSWEVRVIGWEVRYGAEFVPSNEGSYTVIVQK 453
G +D+VTEIT+ PA+KHTVEFPVTE LLSWE+RVIGW+V YGAEFVPS EGSYTVI+QK
Sbjct: 3 GVNDAVTEITVSPAAKHTVEFPVTENGLLSWELRVIGWDVSYGAEFVPSAEGSYTVIIQK 182
Query: 454 ARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKKLLYRLKTK 498
ARKVASSEE VLC ++KI EPGKVVLTIDN SS+KKKLLYRLKTK
Sbjct: 183 ARKVASSEEPVLCQNYKIGEPGKVVLTIDNQSSKKKKLLYRLKTK 317
>TC16991 similar to GB|AAN73306.1|25141223|BT002309 At3g51670/T18N14_50
{Arabidopsis thaliana;}, partial (38%)
Length = 681
Score = 137 bits (346), Expect = 3e-33
Identities = 68/147 (46%), Positives = 101/147 (68%), Gaps = 3/147 (2%)
Frame = +3
Query: 348 SPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKDGEFGNSDS--VTEITIR 405
SPFLTQRTKSKFV + + ETL ++ PE++PV+YGGL + + N +E T++
Sbjct: 3 SPFLTQRTKSKFVISKEGNAAETLYKFVRPEEIPVRYGGLCRPSDLQNGPPKPASEFTVK 182
Query: 406 PASKHTVEFPVTEK-CLLSWEVRVIGWEVRYGAEFVPSNEGSYTVIVQKARKVASSEEAV 464
K ++ E ++W++ V GW++ Y AEFVP+ EGSYT+ V+KARK+ +SEEA+
Sbjct: 183 GGEKVNIQIEGIEYGATITWDIVVGGWDLEYSAEFVPNAEGSYTIAVEKARKIEASEEAI 362
Query: 465 LCNSFKINEPGKVVLTIDNTSSRKKKL 491
NSF + E GK+VL++DN++SRKKK+
Sbjct: 363 -HNSFTLKEAGKMVLSVDNSASRKKKV 440
>BE122488
Length = 346
Score = 107 bits (267), Expect = 5e-24
Identities = 55/110 (50%), Positives = 74/110 (67%), Gaps = 4/110 (3%)
Frame = +2
Query: 325 PEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKY 384
PE VAK +FINVP+WY A+N +ISPFLTQRTKSKFV P+K TETL+ YI E++PV Y
Sbjct: 17 PELVAKNIFINVPFWYYALNALISPFLTQRTKSKFVIGRPAKVTETLIKYIPIEEIPVHY 196
Query: 385 GGLSK--DGEFGNSD-SVTEITIRPASKHTVEFPVTE-KCLLSWEVRVIG 430
GG + D EF D + +E+ ++ S T+E P E + W++ V+G
Sbjct: 197 GGFKREDDSEFTAQDGAASELFLKAGSTATIEIPALEVGSNICWDLTVMG 346
>TC17702 weakly similar to GB|AAN73306.1|25141223|BT002309
At3g51670/T18N14_50 {Arabidopsis thaliana;}, partial
(20%)
Length = 574
Score = 87.0 bits (214), Expect = 6e-18
Identities = 45/83 (54%), Positives = 61/83 (73%), Gaps = 3/83 (3%)
Frame = +2
Query: 166 SIYGIPLL-ADETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLG-DE 223
S++G+ LL D+ +DV+LLKFLRARDF+V +A M+ + WRKEFG + ++DE+LG E
Sbjct: 326 SMWGVTLLNGDDRADVLLLKFLRARDFRVCDALNMLLKCLAWRKEFGADNIVDEELGFKE 505
Query: 224 LEKVV-YMHGFDKEGHPVCYNIY 245
LE VV +D+EGHPVCYN Y
Sbjct: 506 LEGVVACTQVYDREGHPVCYNDY 574
>TC18924 homologue to UP|Q94FN3 (Q94FN3) Phosphatidylinositol transfer-like
protein II, complete
Length = 1976
Score = 75.1 bits (183), Expect = 2e-14
Identities = 59/217 (27%), Positives = 100/217 (45%), Gaps = 12/217 (5%)
Frame = +3
Query: 182 LLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVYM-----HGFDKE 236
LL+FL+AR F +++ M + + WRKEFG + ++++ E+++VV HG DK+
Sbjct: 285 LLRFLKARKFDTEKSKQMWSDMLQWRKEFGADTIVEDFDFKEIDEVVKYYPHGHHGVDKD 464
Query: 237 GHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVN 296
G PV G+ +L T D ++K+ ++ E++ +L F + H++
Sbjct: 465 GRPVYIENIGQVDATKLMQVTTMD-----RYIKYHVKEFERTF-DLKFAACSIAAKKHID 626
Query: 297 D---LKDSPGPGKWELRQATKQALQLFQ----DNYPEFVAKQVFINVPWWYLAVNRMISP 349
+ D G G + ++ + Q DNYPE + + IN + + +
Sbjct: 627 QSTTILDVQGVGLKNFNKHARELITRLQKIDGDNYPETLNRMFIINAGSGFRMLWSTVKS 806
Query: 350 FLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGG 386
FL +T SK G +K LL I QLP GG
Sbjct: 807 FLDPKTTSKIHVLG-NKYQSKLLEVIDASQLPEFLGG 914
>TC13909 weakly similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, partial
(20%)
Length = 399
Score = 71.6 bits (174), Expect = 3e-13
Identities = 32/50 (64%), Positives = 42/50 (84%)
Frame = +3
Query: 163 EQVSIYGIPLLADETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFGI 212
++ SI+G+PLL D+ +DVILLKFLRAR+FKVK+A MI NT+ WRK+F I
Sbjct: 249 DEPSIWGVPLLKDDRTDVILLKFLRAREFKVKDALLMINNTLRWRKDFNI 398
>TC8616 UP|Q94FN1 (Q94FN1) Phosphatidylinositol transfer-like protein III,
complete
Length = 2302
Score = 52.0 bits (123), Expect = 2e-07
Identities = 27/65 (41%), Positives = 40/65 (61%), Gaps = 5/65 (7%)
Frame = +2
Query: 181 ILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVYM-----HGFDK 235
++L+FL+AR F +++A M + WRKEFG + +M + EL++VV HG DK
Sbjct: 404 MMLRFLKARKFDIEKAKHMWAEMLQWRKEFGADTIMQDFEFQELDEVVRYYPHGHHGVDK 583
Query: 236 EGHPV 240
EG PV
Sbjct: 584 EGRPV 598
>AV408295
Length = 426
Score = 45.1 bits (105), Expect = 3e-05
Identities = 34/111 (30%), Positives = 50/111 (44%), Gaps = 12/111 (10%)
Frame = +3
Query: 56 NIPNNENNSLQELQNLIQQAFNNHAFSAPPLIKEQKQSTTT---TVAAEPAQENKYQLED 112
++P + +L +L+ LIQ A N H SAPP K ST VAA +E + E+
Sbjct: 27 DLPEPQKKALDDLKQLIQDALNKHELSAPPPPKAPSPSTEAKKDEVAAAATEEKVEEEEE 206
Query: 113 KK---------ENVVSSVEDDGAKTVEAIEESIVAVSASVPPEQKPVVEKV 154
KK E V ++ K E +++ A+ E+K VVE V
Sbjct: 207 KKTEEEKKPAEEEKKEEVVEEEKKPAEEEKKAEAEGEATTSGEEKNVVEAV 359
>TC11997 similar to UP|O48940 (O48940) Polyphosphoinositide binding protein
Ssh2p, partial (26%)
Length = 492
Score = 43.5 bits (101), Expect = 8e-05
Identities = 21/57 (36%), Positives = 30/57 (51%)
Frame = +2
Query: 330 KQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGG 386
K ++ P+ ++ V +++ PF+ TK K VF K TLL I QLP YGG
Sbjct: 2 KLFIVHAPYIFMKVWKLVYPFIDDHTKKKIVFVDNQKLKSTLLEEIDESQLPEIYGG 172
>TC18426
Length = 357
Score = 39.3 bits (90), Expect = 0.002
Identities = 23/66 (34%), Positives = 40/66 (59%), Gaps = 7/66 (10%)
Frame = +3
Query: 24 PQPQKKNNPVTLQSHLSKPNTEQEQ----TFNKPSD---NIPNNENNSLQELQNLIQQAF 76
P+P+++ PV ++ +SKP E +F + S+ ++P+ E+ +LQE + L+Q A
Sbjct: 162 PKPEEEK-PVEAETEVSKPGGGDEVPESGSFKEESNIVADLPDAESKALQEFKLLVQTAL 338
Query: 77 NNHAFS 82
NNH FS
Sbjct: 339 NNHEFS 356
>TC16241 weakly similar to UP|Q91TH4 (Q91TH4) T122, partial (7%)
Length = 799
Score = 36.2 bits (82), Expect = 0.013
Identities = 31/142 (21%), Positives = 51/142 (35%)
Frame = +1
Query: 18 QPLYSHPQPQKKNNPVTLQSHLSKPNTEQEQTFNKPSDNIPNNENNSLQELQNLIQQAFN 77
QP Y+ P P K +P + +++ P T P+ P N + + L +
Sbjct: 10 QPPYT-PAPIKPPSPTSQPPYIATPPNTHSPTSQPPNTQTPANTPSPSSQPPYLSTPPKS 186
Query: 78 NHAFSAPPLIKEQKQSTTTTVAAEPAQENKYQLEDKKENVVSSVEDDGAKTVEAIEESIV 137
N S PP+ + + T + + N ++ S+ A S
Sbjct: 187 NSPTSQPPVAQTPRAVTPISSPSSSPTSNSPYPSTNPPSLAPSISTSPPALTPA---STP 357
Query: 138 AVSASVPPEQKPVVEKVEASLP 159
+ + PP Q P E ASLP
Sbjct: 358 VPATASPPSQSPSAESPGASLP 423
>TC14873 weakly similar to GB|BAB08982.1|9758453|AB008271 selenium-binding
protein-like {Arabidopsis thaliana;} , partial (7%)
Length = 763
Score = 34.3 bits (77), Expect = 0.049
Identities = 21/66 (31%), Positives = 32/66 (47%), Gaps = 2/66 (3%)
Frame = +2
Query: 323 NYPEFVAKQVFINVPWWYLAVNRMISPFLTQRT--KSKFVFAGPSKSTETLLSYIAPEQL 380
+YPE +A N P + A + + FL +T K KFV+ S E + S + L
Sbjct: 80 HYPERLAIAFLFNPPSIFQAFYKAVKYFLDPKTAQKVKFVYPNNKDSVELMKSLFDIDNL 259
Query: 381 PVKYGG 386
P ++GG
Sbjct: 260 PSEFGG 277
>TC9914 weakly similar to GB|AAN73306.1|25141223|BT002309
At3g51670/T18N14_50 {Arabidopsis thaliana;}, partial
(7%)
Length = 469
Score = 33.5 bits (75), Expect = 0.083
Identities = 14/24 (58%), Positives = 19/24 (78%)
Frame = +1
Query: 177 TSDVILLKFLRARDFKVKEAFTMI 200
T+DVILLK +R RDF+V EA ++
Sbjct: 133 TADVILLKIIRTRDFRVSEALNIL 204
>TC18777 weakly similar to UP|Q9LDJ7 (Q9LDJ7) ESTs AU081555, partial (24%)
Length = 693
Score = 32.7 bits (73), Expect = 0.14
Identities = 17/80 (21%), Positives = 34/80 (42%), Gaps = 2/80 (2%)
Frame = +1
Query: 314 KQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQR--TKSKFVFAGPSKSTETL 371
++ + + Q+ YPE + + N PW + M+ P L K KF ++ + + +
Sbjct: 40 RETIHVLQEYYPERLGLSLIYNAPWIFQPFLAMVKPLLQPENYNKLKFGYSNDEGTKKIM 219
Query: 372 LSYIAPEQLPVKYGGLSKDG 391
+ L +GG + G
Sbjct: 220 EDLFDMDHLESAFGGNNNTG 279
>AV428489
Length = 386
Score = 32.0 bits (71), Expect = 0.24
Identities = 20/60 (33%), Positives = 29/60 (48%), Gaps = 1/60 (1%)
Frame = +1
Query: 323 NYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKF-VFAGPSKSTETLLSYIAPEQLP 381
NYPE +N P+ + A +++ P L +RT+ K V G K E LL + LP
Sbjct: 103 NYPEKTDTYYIVNAPYVFSACWKVVKPLLQERTRRKIQVLQGSGK--EELLKIMDYASLP 276
>TC8445 similar to UP|O80910 (O80910) At2g38410 protein, partial (7%)
Length = 841
Score = 31.6 bits (70), Expect = 0.32
Identities = 40/159 (25%), Positives = 57/159 (35%), Gaps = 15/159 (9%)
Frame = +1
Query: 8 NQTTPHVYVPQP-----------LYSHPQPQKKNNPVTLQSHLSKPNTE-QEQTFNKPSD 55
N+ P Y PQP HPQPQ ++ P S+P + Q Q +P
Sbjct: 19 NEPQPLQYQPQPHLQHQPQHHPHFQPHPQPQLQSQPQPQHIQQSQPQPQSQPQHIQQPQ- 195
Query: 56 NIPNNENNSLQELQNLIQQAFNNHAFSAPPLIKEQKQSTTTTVAAEPAQENKYQLEDKKE 115
P + +Q+ Q Q F N F P AA P N YQ
Sbjct: 196 --PQPQLQHIQQSQP--QPQFQNQHFQYPGTYPPPPW------AATPGYPN-YQNHLSST 342
Query: 116 NVVSSVEDDGAKTVEAIEESIVAVSASVPPE---QKPVV 151
N +S+ + + T + + + S PP Q+P V
Sbjct: 343 NALSTPQANTPYTSAVRGDPVASSGQSNPPNATGQRPFV 459
>TC12614
Length = 577
Score = 31.2 bits (69), Expect = 0.41
Identities = 30/94 (31%), Positives = 43/94 (44%), Gaps = 3/94 (3%)
Frame = +1
Query: 145 PEQKPVVEKVEASLPLPPEQVSIYGIP---LLADETSDVILLKFLRARDFKVKEAFTMIK 201
P +KP + + L P Q S+ P A+ SD+ LR RD K E
Sbjct: 184 PLKKPKMNNLLLPLTEPQHQHSLAFEPDTAAAAEPDSDISYGLNLR-RDAKQTEDNAHDD 360
Query: 202 NTILWRKEFGIEELMDEKLGDELEKVVYMHGFDK 235
+ R+ F E +M +KL D+LEK+ GFD+
Sbjct: 361 DAAAPRRRFQPEAMMLQKLKDDLEKLPDDQGFDE 462
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.313 0.131 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,436,857
Number of Sequences: 28460
Number of extensions: 117576
Number of successful extensions: 616
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 599
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 606
length of query: 503
length of database: 4,897,600
effective HSP length: 94
effective length of query: 409
effective length of database: 2,222,360
effective search space: 908945240
effective search space used: 908945240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC148755.7


