

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148755.4 + phase: 1 /partial
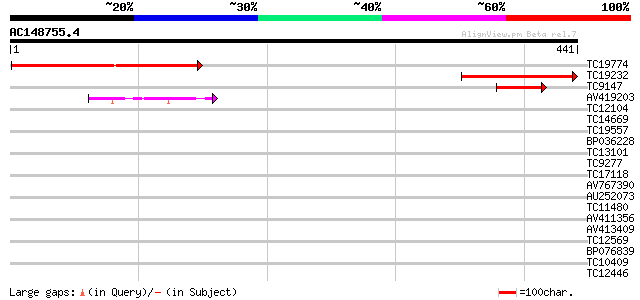
(441 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC19774 weakly similar to UP|AAR92268 (AAR92268) At1g47570, part... 203 4e-53
TC19232 similar to UP|AAR92268 (AAR92268) At1g47570, partial (19%) 164 2e-41
TC9147 similar to UP|AAR92268 (AAR92268) At1g47570, partial (8%) 81 3e-16
AV419203 45 3e-05
TC12104 40 0.001
TC14669 similar to PIR|E86326|E86326 protein F18O14.3 [imported]... 39 0.001
TC19557 similar to PIR|E86326|E86326 protein F18O14.3 [imported]... 39 0.002
BP036228 39 0.002
TC13101 similar to PIR|E86378|E86378 protein F21J9.10 [imported]... 38 0.004
TC9277 similar to GB|AAP13413.1|30023760|BT006305 At3g55530 {Ara... 37 0.005
TC17118 similar to UP|Q94CL1 (Q94CL1) RING finger-like protein (... 37 0.006
AV767390 35 0.019
AU252073 35 0.025
TC11480 similar to UP|Q8VYW5 (Q8VYW5) AT5g45290/K9E15_7, partial... 35 0.032
AV411356 35 0.032
AV413409 35 0.032
TC12569 UP|Q40221 (Q40221) Protein containing C-terminal RING-fi... 34 0.042
BP076839 33 0.072
TC10409 similar to UP|Q8GUU2 (Q8GUU2) RES protein, partial (17%) 33 0.072
TC12446 similar to PIR|T51854|T51854 RING-H2 finger protein RHF2... 33 0.12
>TC19774 weakly similar to UP|AAR92268 (AAR92268) At1g47570, partial (19%)
Length = 642
Score = 203 bits (517), Expect = 4e-53
Identities = 102/149 (68%), Positives = 112/149 (74%)
Frame = +2
Query: 2 SDSRYPDVEIRSDKGQIRSEISTASCDKHRWCKIVRNSDLCSATLKNKSSNTILVDGAEV 61
SDSRY D+ I SD I SEIS K WCKI RNSDLCSAT++N S NTILVDGA V
Sbjct: 194 SDSRYSDIVIWSDVTGICSEISATPSVKRCWCKIARNSDLCSATMENTSPNTILVDGAVV 373
Query: 62 GKGDTVVIKDRSEIIPGPAKEGFVSYKFQIVSSPEICQTLLKICVDVDHAKCSICLNIWH 121
+T+ IKD SEII G K G+VSY+F I+SSPE CQ LKICVDV++ KCSICLNIWH
Sbjct: 374 CNDETIAIKDGSEIISGSDK-GYVSYRFHIMSSPETCQRELKICVDVENTKCSICLNIWH 550
Query: 122 DVVTVAPCLHNFCNGCFSEWLRRSQEKRS 150
DVVT PCLHNFCNGCFSEW RRSQ K S
Sbjct: 551 DVVTTGPCLHNFCNGCFSEWSRRSQAKCS 637
>TC19232 similar to UP|AAR92268 (AAR92268) At1g47570, partial (19%)
Length = 477
Score = 164 bits (416), Expect = 2e-41
Identities = 73/90 (81%), Positives = 78/90 (86%)
Frame = +2
Query: 352 LNNREIDTTRLMLNHAEMMTAGTLVCSDCYQKLVSFFLYWFRISTPKHLLPPEASTREDC 411
L N EID TRLMLNH + +T+ T VC DCYQKLVSF LYWFRIS P+ LLPP+AS REDC
Sbjct: 2 LYNGEIDRTRLMLNHVDTITSRTFVCGDCYQKLVSFLLYWFRISIPQRLLPPDASAREDC 181
Query: 412 WYGYACRTQHRSEEHARKRNHVCRPTRGSH 441
WYGYACRTQHRSEEHARKRNHVCRPTRGSH
Sbjct: 182 WYGYACRTQHRSEEHARKRNHVCRPTRGSH 271
>TC9147 similar to UP|AAR92268 (AAR92268) At1g47570, partial (8%)
Length = 657
Score = 81.3 bits (199), Expect = 3e-16
Identities = 33/39 (84%), Positives = 35/39 (89%)
Frame = +1
Query: 379 DCYQKLVSFFLYWFRISTPKHLLPPEASTREDCWYGYAC 417
DCYQKLVSF LYWFRIS P+ LLPP+AS REDCWYGYAC
Sbjct: 253 DCYQKLVSFLLYWFRISIPQRLLPPDASAREDCWYGYAC 369
>AV419203
Length = 393
Score = 44.7 bits (104), Expect = 3e-05
Identities = 35/106 (33%), Positives = 49/106 (46%), Gaps = 6/106 (5%)
Frame = +1
Query: 62 GKGDTVVIKDRSEIIPG----PAKEGFVSYKFQIVSSPEICQTLLKICVDVDHAKCSICL 117
G+ V+ +D + PG PA+ V Q E+ + LK V D ++C +CL
Sbjct: 109 GQETRVMAQDAATYQPGLYLTPAQREAVEALIQ-----ELPKFRLK-AVPTDCSECPVCL 270
Query: 118 NIWHD--VVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVV 161
+H+ V PC HNF C EWLR + V CP+CR V
Sbjct: 271 EEFHEGNEVRGLPCAHNFHVECIDEWLRLN------VKCPRCRCSV 390
>TC12104
Length = 771
Score = 39.7 bits (91), Expect = 0.001
Identities = 19/50 (38%), Positives = 24/50 (48%)
Frame = +1
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQ 162
C+IC H + + C H FC C SEW R + CP CRA V+
Sbjct: 421 CAICQEKMHSPILLC-CKHMFCEECVSEWFERER------TCPLCRASVK 549
>TC14669 similar to PIR|E86326|E86326 protein F18O14.3 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(77%)
Length = 1120
Score = 39.3 bits (90), Expect = 0.001
Identities = 18/56 (32%), Positives = 30/56 (53%)
Frame = +2
Query: 107 DVDHAKCSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQ 162
D + +C+IC ++ D + + C H FC C +WL + R CP C+A+V+
Sbjct: 110 DAGNFECNICFDLAQDPI-ITLCGHLFCWPCLYKWLHFHSQSRE---CPVCKALVE 265
>TC19557 similar to PIR|E86326|E86326 protein F18O14.3 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(24%)
Length = 457
Score = 38.5 bits (88), Expect = 0.002
Identities = 18/56 (32%), Positives = 27/56 (48%)
Frame = +2
Query: 107 DVDHAKCSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQ 162
D +C+IC + D V + C H FC C WL + + CP C+A++Q
Sbjct: 293 DAGDFECNICFELAQDPV-ITLCGHLFCWPCLYRWLHHHSQSQE---CPVCKALIQ 448
>BP036228
Length = 626
Score = 38.5 bits (88), Expect = 0.002
Identities = 20/46 (43%), Positives = 25/46 (53%)
Frame = +2
Query: 113 CSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCR 158
CS C+ + VT PC HNFC CF +W+ Q KR+ C CR
Sbjct: 422 CSFCMQLPDRPVTT-PCGHNFCLKCFEKWI--GQGKRT---CANCR 541
>TC13101 similar to PIR|E86378|E86378 protein F21J9.10 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(47%)
Length = 588
Score = 37.7 bits (86), Expect = 0.004
Identities = 23/75 (30%), Positives = 40/75 (52%), Gaps = 1/75 (1%)
Frame = +3
Query: 112 KCSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFV-GKNHFL 170
+C ICL +V + C H C C+ +W R+S+ CP CR+ ++ V ++ ++
Sbjct: 153 ECGICLEPCTKIV-LPYCCHAMCIKCYCKWNRKSES------CPFCRSSLRRVKSEDLWV 311
Query: 171 RTIAEDMVKADSSLR 185
T ED+V A++ R
Sbjct: 312 LTCNEDVVDAETVSR 356
>TC9277 similar to GB|AAP13413.1|30023760|BT006305 At3g55530 {Arabidopsis
thaliana;}, partial (79%)
Length = 1297
Score = 37.4 bits (85), Expect = 0.005
Identities = 19/55 (34%), Positives = 25/55 (44%), Gaps = 2/55 (3%)
Frame = +3
Query: 106 VDVDHAKCSICLNIWH--DVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCR 158
V D CS+CL + D++ PCLH F C WLR+ CP C+
Sbjct: 861 VSDDELTCSVCLEQVNVGDILRSLPCLHQFHANCIDPWLRQQG------TCPVCK 1007
>TC17118 similar to UP|Q94CL1 (Q94CL1) RING finger-like protein
(AT5g19430/F7K24_180), partial (35%)
Length = 708
Score = 37.0 bits (84), Expect = 0.006
Identities = 20/72 (27%), Positives = 33/72 (45%), Gaps = 2/72 (2%)
Frame = +2
Query: 113 CSICLN--IWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFVGKNHFL 170
C+ICL+ + + V C H +C C W + + V CPQC+ +F+ + L
Sbjct: 341 CAICLDKIVLQETALVKGCEHAYCVTCILRWATYNNK----VTCPQCKNPFEFLNVHRSL 508
Query: 171 RTIAEDMVKADS 182
ED + +S
Sbjct: 509 DGSIEDYMFEES 544
>AV767390
Length = 447
Score = 35.4 bits (80), Expect = 0.019
Identities = 18/52 (34%), Positives = 26/52 (49%), Gaps = 2/52 (3%)
Frame = -3
Query: 109 DHAKCSICLNIWHDVVTVA--PCLHNFCNGCFSEWLRRSQEKRSTVLCPQCR 158
+ A+C ICL+ + D V + PC H+F GC +WL CP C+
Sbjct: 442 EDAECCICLSSYDDGVELRELPCGHHFHCGCVDKWL------HINATCPLCK 305
>AU252073
Length = 258
Score = 35.0 bits (79), Expect = 0.025
Identities = 26/86 (30%), Positives = 42/86 (48%), Gaps = 3/86 (3%)
Frame = +1
Query: 108 VDHAKCSICLNIW--HDVVTVAP-CLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVVQFV 164
++ +C ICL + +V+ + P C H F C WLR K+ST CP CR ++
Sbjct: 19 IEDTQCVICLADYKEREVLRIMPKCGHTFHLSCIDIWLR----KQST--CPVCRLPLKNS 180
Query: 165 GKNHFLRTIAEDMVKADSSLRRSHDE 190
+ +R V +++ +SHDE
Sbjct: 181 SETKHVRP-----VTFTTTMSQSHDE 243
>TC11480 similar to UP|Q8VYW5 (Q8VYW5) AT5g45290/K9E15_7, partial (8%)
Length = 502
Score = 34.7 bits (78), Expect = 0.032
Identities = 20/57 (35%), Positives = 26/57 (45%), Gaps = 2/57 (3%)
Frame = +2
Query: 107 DVDHAKCSICLNIWH--DVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVV 161
D D +C ICL + D + V PC H + C +WL K +CP CR V
Sbjct: 104 DHDAEQCHICLAEYEEGDQIRVLPCNHEYHKLCVDKWL-----KEIHGVCPLCRGNV 259
>AV411356
Length = 358
Score = 34.7 bits (78), Expect = 0.032
Identities = 19/55 (34%), Positives = 26/55 (46%)
Frame = +2
Query: 107 DVDHAKCSICLNIWHDVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVV 161
D D C+ICL++ + V V C H FC C WL + + CP C+ V
Sbjct: 26 DGDFFDCNICLDLAREPV-VTCCGHLFCWTCVYRWLHLHSDAKE---CPVCKGEV 178
>AV413409
Length = 374
Score = 34.7 bits (78), Expect = 0.032
Identities = 19/51 (37%), Positives = 24/51 (46%), Gaps = 3/51 (5%)
Frame = +1
Query: 112 KCSICLNIWHDVVT---VAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRA 159
+C++CLN + D T + C H F C EWL S CP CRA
Sbjct: 46 ECAVCLNEFEDTETLRLIPKCDHVFHPECIDEWL------SSHSTCPVCRA 180
>TC12569 UP|Q40221 (Q40221) Protein containing C-terminal RING-finger,
complete
Length = 2092
Score = 34.3 bits (77), Expect = 0.042
Identities = 19/56 (33%), Positives = 26/56 (45%), Gaps = 3/56 (5%)
Frame = +2
Query: 109 DHAKCSICLNIWH---DVVTVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQCRAVV 161
D C ICL + DV T+ C H++ C +WL + LCP C+A V
Sbjct: 1772 DEGSCVICLEEYKNMDDVGTLKTCGHDYHVNCIKKWLSMKK------LCPICKASV 1921
>BP076839
Length = 392
Score = 33.5 bits (75), Expect = 0.072
Identities = 17/55 (30%), Positives = 27/55 (48%), Gaps = 2/55 (3%)
Frame = -3
Query: 106 VDVDHAKCSICLNIWHDVVTVA--PCLHNFCNGCFSEWLRRSQEKRSTVLCPQCR 158
+ + A+C ICL+ + D V + PC H+F C +WL + CP C+
Sbjct: 381 LSAEDAECCICLSAYDDGVELRQLPCGHHFHCACVDKWLCMN------ATCPLCK 235
>TC10409 similar to UP|Q8GUU2 (Q8GUU2) RES protein, partial (17%)
Length = 715
Score = 33.5 bits (75), Expect = 0.072
Identities = 15/52 (28%), Positives = 27/52 (51%), Gaps = 2/52 (3%)
Frame = +3
Query: 109 DHAKCSICLNIWHDVVTV--APCLHNFCNGCFSEWLRRSQEKRSTVLCPQCR 158
+ A+C IC++ + D + PC H+F + C +WL+ + CP C+
Sbjct: 33 EDAECCICISSYEDGAELHSLPCNHHFHSTCIVKWLKMN------ATCPLCK 170
>TC12446 similar to PIR|T51854|T51854 RING-H2 finger protein RHF2a
[imported] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (38%)
Length = 550
Score = 32.7 bits (73), Expect = 0.12
Identities = 19/47 (40%), Positives = 21/47 (44%), Gaps = 2/47 (4%)
Frame = +2
Query: 113 CSICLNIWHDVV--TVAPCLHNFCNGCFSEWLRRSQEKRSTVLCPQC 157
CSICL D TV C H F C EW +RS + CP C
Sbjct: 203 CSICLEDLCDSEPSTVTGCKHEFHLQCILEWCQRSSQ------CPMC 325
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.134 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,596,413
Number of Sequences: 28460
Number of extensions: 168195
Number of successful extensions: 885
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 880
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 884
length of query: 441
length of database: 4,897,600
effective HSP length: 93
effective length of query: 348
effective length of database: 2,250,820
effective search space: 783285360
effective search space used: 783285360
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC148755.4


