

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148718.8 - phase: 0
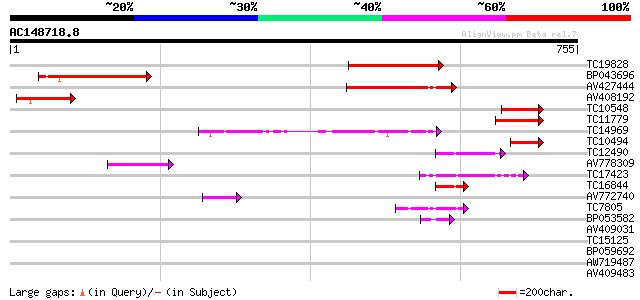
(755 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC19828 similar to UP|Q9FNT6 (Q9FNT6) Long chain acyl-CoA synthe... 211 5e-55
BP043696 159 2e-39
AV427444 130 6e-31
AV408192 124 6e-29
TC10548 similar to UP|Q940V0 (Q940V0) T23O15.3/T23O15.3, partial... 98 6e-21
TC11779 similar to UP|Q9FNT6 (Q9FNT6) Long chain acyl-CoA synthe... 85 5e-17
TC14969 similar to PIR|PQ0772|PQ0772 4-coumarate-CoA ligase (cl... 69 3e-12
TC10494 similar to UP|Q9FNT6 (Q9FNT6) Long chain acyl-CoA synthe... 62 3e-10
TC12490 similar to UP|Q9FFE6 (Q9FFE6) AMP-binding protein (Adeno... 53 2e-07
AV778309 50 1e-06
TC17423 similar to PIR|H85064|H85064 4-coumarate-CoA ligase-like... 45 6e-05
TC16844 similar to UP|Q8LRT6 (Q8LRT6) Adenosine monophosphate bi... 45 6e-05
AV772740 44 7e-05
TC7805 homologue to PIR|PQ0772|PQ0772 4-coumarate-CoA ligase (c... 44 1e-04
BP053582 42 5e-04
AV409031 38 0.005
TC15125 weakly similar to UP|Q84P20 (Q84P20) Acyl-activating enz... 37 0.009
BP059692 37 0.009
AW719487 37 0.009
AV409483 33 0.13
>TC19828 similar to UP|Q9FNT6 (Q9FNT6) Long chain acyl-CoA synthetase ,
partial (17%)
Length = 378
Score = 211 bits (536), Expect = 5e-55
Identities = 94/126 (74%), Positives = 111/126 (87%)
Frame = +1
Query: 452 NLRFMLCGGAPLSGDSQQFINICVGAPIGQGYGLTETFAGAAFSEADDYSVGRVGPPLPC 511
++RF+L GGAPLS D+Q+FINIC+GAPIGQGYGLTET AG FS+ DD SVGRVGPPLPC
Sbjct: 1 HIRFILSGGAPLSADTQRFINICLGAPIGQGYGLTETCAGGTFSDFDDTSVGRVGPPLPC 180
Query: 512 CYIKLVSWEEGGYLSSDKPMPRGEVVVGGFSVTAGYFKNQDKTDEVFKVDEKGVRWFYTG 571
YIKL+ W EGGY ++D PMPRGE+VVGG +VT GYFKN++KT+E +KVDE+GVRWFYTG
Sbjct: 181 SYIKLIDWPEGGYSTNDSPMPRGEIVVGGPNVTLGYFKNEEKTNESYKVDERGVRWFYTG 360
Query: 572 DIGRFH 577
DIGRFH
Sbjct: 361 DIGRFH 378
>BP043696
Length = 470
Score = 159 bits (402), Expect = 2e-39
Identities = 81/155 (52%), Positives = 103/155 (66%), Gaps = 5/155 (3%)
Frame = +1
Query: 39 IVPVLFSVFFLGKKKGKVRGVPVEVSG-----ESGYAVRNARYSELVEVPWKGAPTMAHL 93
+ P+L S+ F +K K RGV ++ + ESG VRNAR+ V W+G T+A L
Sbjct: 10 LFPILASLLF--RKPPKHRGVTLDTTATTDDSESGVTVRNARFDAPVVTAWEGVTTLAEL 183
Query: 94 FEPSCKKHTHNRFLGTRKLIGKEFVTSSDGRKFEKVHLGEYEWETYGEVFARVSNFASGL 153
FE +C+KH LG R++I KE SDGR FEKVHLGEY W +Y VF V++FASGL
Sbjct: 184 FEEACRKHGERALLGAREVISKEAEAGSDGRVFEKVHLGEYRWLSYDAVFEAVTSFASGL 363
Query: 154 LKLGHDIDSHVAIFSDTRAEWFIALQGCFRQNITV 188
+LGH + VAIF+DTR EWFIALQGCFR+N+TV
Sbjct: 364 KELGHGREERVAIFADTREEWFIALQGCFRRNVTV 468
>AV427444
Length = 428
Score = 130 bits (328), Expect = 6e-31
Identities = 68/146 (46%), Positives = 92/146 (62%)
Frame = +1
Query: 449 LGGNLRFMLCGGAPLSGDSQQFINICVGAPIGQGYGLTETFAGAAFSEADDYSVGRVGPP 508
LGG +R M+ G +PLS D +F+ IC G + +GYG+TET + + D G VG P
Sbjct: 1 LGGRVRLMVSGASPLSPDIMEFLKICFGGRVTEGYGMTETTCVISCIDQGDKLGGHVGSP 180
Query: 509 LPCCYIKLVSWEEGGYLSSDKPMPRGEVVVGGFSVTAGYFKNQDKTDEVFKVDEKGVRWF 568
P C +KLV E Y S D+P PRGE+ V G + GY+K++ +T EV +DE+G W
Sbjct: 181 SPACEVKLVDVPEMNYTSDDQPQPRGEICVRGPIIFKGYYKDEAQTREV--IDEEG--WL 348
Query: 569 YTGDIGRFHPDGCLEIIDRKKDIVKL 594
+TGDIG + P G L+IIDRKK+I KL
Sbjct: 349 HTGDIGTWLPGGRLKIIDRKKNIFKL 426
>AV408192
Length = 386
Score = 124 bits (311), Expect = 6e-29
Identities = 63/85 (74%), Positives = 70/85 (82%), Gaps = 6/85 (7%)
Frame = +1
Query: 9 SWLKSLNICETFGLKDHG------ELGTVFAIVIGIIVPVLFSVFFLGKKKGKVRGVPVE 62
SWLKSLNI + FGLKDHG E G V AI IGI+ PVLFSV FLGKKKGKVRGVPV+
Sbjct: 130 SWLKSLNISDLFGLKDHGHGHGHGEYGAVAAIFIGILFPVLFSVLFLGKKKGKVRGVPVQ 309
Query: 63 VSGESGYAVRNARYSELVEVPWKGA 87
VSGE+GYAVRNAR +ELV+VPW+GA
Sbjct: 310 VSGEAGYAVRNARKTELVQVPWEGA 384
>TC10548 similar to UP|Q940V0 (Q940V0) T23O15.3/T23O15.3, partial (10%)
Length = 760
Score = 97.8 bits (242), Expect = 6e-21
Identities = 47/55 (85%), Positives = 51/55 (92%)
Frame = +3
Query: 656 DLCNKPEAVTEVLQSISKAAKAAKLQKTEVPAKIKLLADPWTPESGLVTAALKLK 710
DLCNKPE VTEVLQSISK AKA+KL+K+E+P KIKLL DPWTPESGLVTAALKLK
Sbjct: 3 DLCNKPETVTEVLQSISKVAKASKLEKSEIPKKIKLLPDPWTPESGLVTAALKLK 167
>TC11779 similar to UP|Q9FNT6 (Q9FNT6) Long chain acyl-CoA synthetase ,
partial (11%)
Length = 612
Score = 84.7 bits (208), Expect = 5e-17
Identities = 41/63 (65%), Positives = 49/63 (77%)
Frame = +2
Query: 648 GIEYKDFPDLCNKPEAVTEVLQSISKAAKAAKLQKTEVPAKIKLLADPWTPESGLVTAAL 707
GI Y D +LC K E V EV S+ K AK A+L+K E+PAKIKLL+DPW+PESGLVTAAL
Sbjct: 62 GISYFDLSELCTKEETVKEVHASLVKEAKKARLEKFEIPAKIKLLSDPWSPESGLVTAAL 241
Query: 708 KLK 710
K+K
Sbjct: 242 KIK 250
>TC14969 similar to PIR|PQ0772|PQ0772 4-coumarate-CoA ligase (clone
GM4CL1B) - soybean (fragment) {Glycine
max;} , partial (70%)
Length = 928
Score = 68.9 bits (167), Expect = 3e-12
Identities = 91/344 (26%), Positives = 148/344 (42%), Gaps = 21/344 (6%)
Frame = +2
Query: 252 LSSNCKIASFDEVEK---------LGKESPV-EPSLPSKNAVAVVMYTSGSTGLPKGVMI 301
L + K+ + DE K GKE + E + ++AVA+ ++SG+TGLPKGV++
Sbjct: 80 LGEDFKVVTVDEPPKDCLDFSVISEGKEDDLPEVEINPEDAVALP-FSSGTTGLPKGVVL 256
Query: 302 THGNIVATTAA-VMTVIPNL--GSKDVYLAYLPLAHVFEMAAESVMLAAGVAIGYGSPMT 358
TH ++ + V PNL G++DV L LPL H+F + SV+L A+ GS +
Sbjct: 257 THKSLTTSVGQQVDGENPNLSLGTEDVLLCVLPLFHIFSL--NSVLLC---ALRAGSGVL 421
Query: 359 LTDTSNKVKKGTKGDVTVLKPTLLTAVPAIIDRIRDGVVKKVEEKGGLAKNLFQIAYKRR 418
L K + G+ L ++ K R
Sbjct: 422 LMP---KFEIGS---------------------------------------LLELIQKHR 475
Query: 419 LAAVKGSWLGAWGVEKLVWDTIIFKKIRTVLGGNLRFMLCGGAPLSGDSQQFINICVG-A 477
++ A V LV K+ ++R +L G APL + ++ + V A
Sbjct: 476 VSV-------AMVVPPLVLALAKNPKVAEFDLSSIRLVLSGAAPLGKELEEALRSRVPQA 634
Query: 478 PIGQGYGLTETFAGAAFSEADDYS-------VGRVGPPLPCCYIKLVSWEEGGYLSSDKP 530
+GQGYG+TE AG S + ++ G G + +K++ E G L ++P
Sbjct: 635 VLGQGYGMTE--AGPVLSMSLGFAKQPFPTKSGSCGTVVRNAELKVLDPETGCSLGYNQP 808
Query: 531 MPRGEVVVGGFSVTAGYFKNQDKTDEVFKVDEKGVRWFYTGDIG 574
GE+ + G + GY ++ T +D +G W +TGD+G
Sbjct: 809 ---GEICIRGQQIMKGYLNDEKAT--ATTIDAEG--WLHTGDVG 919
>TC10494 similar to UP|Q9FNT6 (Q9FNT6) Long chain acyl-CoA synthetase ,
partial (7%)
Length = 506
Score = 62.4 bits (150), Expect = 3e-10
Identities = 29/44 (65%), Positives = 38/44 (85%)
Frame = +3
Query: 667 VLQSISKAAKAAKLQKTEVPAKIKLLADPWTPESGLVTAALKLK 710
+L + + AK A+L+K E+PAKIK+L+DPWTPE+GLVTAALKLK
Sbjct: 75 ILGNHLQEAKKARLEKFEIPAKIKVLSDPWTPETGLVTAALKLK 206
>TC12490 similar to UP|Q9FFE6 (Q9FFE6) AMP-binding protein (Adenosine
monophosphate binding protein 5 AMPBP5), partial (21%)
Length = 387
Score = 52.8 bits (125), Expect = 2e-07
Identities = 32/96 (33%), Positives = 50/96 (51%), Gaps = 2/96 (2%)
Frame = +3
Query: 567 WFYTGDIGRFHPDGCLEIIDRKKDIVKLQHGEYISLGKVEAALASGDYVD--SIMVHADP 624
WFYTGD+G H DG LEI DR KD++ + GE +S +VE+ L V+ +++ AD
Sbjct: 36 WFYTGDVGVMHEDGYLEIKDRSKDVI-ISGGENLSTVEVESVLYGHPAVNQAAVVARADE 212
Query: 625 FHSYCVALVVASHQSLEKWAQETGIEYKDFPDLCNK 660
F V+ + L++ E ++ + C K
Sbjct: 213 FWGETPCAFVSLKEGLKETVTE-----REVVEFCRK 305
>AV778309
Length = 543
Score = 50.4 bits (119), Expect = 1e-06
Identities = 26/89 (29%), Positives = 42/89 (46%), Gaps = 1/89 (1%)
Frame = +2
Query: 131 LGEYEWETYGEVFARVSNFASGLLKLGHDIDSHVAIFSDTRAEWFIALQGCFRQNITVVT 190
+G YEW TY E + S + G + I+ EW +A++ C +T V
Sbjct: 236 VGAYEWVTYKEAYDAAIRMGSAMNSCGVNPGDRCGIYGSNCPEWIMAMEACNSYGVTYVP 415
Query: 191 IYASLGEDALIHSLNETQVS-TLICDVKL 218
+Y +LG A+ +N +VS L+ D K+
Sbjct: 416 LYDTLGPKAVEFIINHAEVSIALVQDSKI 502
>TC17423 similar to PIR|H85064|H85064 4-coumarate-CoA ligase-like protein
[imported] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (25%)
Length = 655
Score = 44.7 bits (104), Expect = 6e-05
Identities = 50/150 (33%), Positives = 76/150 (50%), Gaps = 5/150 (3%)
Frame = +1
Query: 546 GYFKNQDKTDEVFKVDEKGVRWFYTGDIGRFHPDGCLEIIDRKKDIVKLQHGEYISLGKV 605
GY N + T +D+KG W +TGD+G F DG L ++DR K+++K + G ++ ++
Sbjct: 16 GYHNNPEATK--LTLDKKG--WVHTGDVGYFDEDGQLYVVDRIKELIKYK-GFQVAPAEL 180
Query: 606 EAALAS-GDYVDSIMV--HADPFHSYCVALVVASHQS--LEKWAQETGIEYKDFPDLCNK 660
E L S + +D+ +V D +A VV S S EK QE I + P K
Sbjct: 181 EGLLVSHPEILDAAVVPYPDDEAGEVPIAYVVRSPNSSLTEKQIQEF-IASQVAP--FKK 351
Query: 661 PEAVTEVLQSISKAAKAAKLQKTEVPAKIK 690
VT + S+ K A + KL + E+ AK K
Sbjct: 352 LRRVT-FISSVPKTA-SGKLLRRELIAKAK 435
>TC16844 similar to UP|Q8LRT6 (Q8LRT6) Adenosine monophosphate binding
protein 1 AMPBP1, partial (25%)
Length = 663
Score = 44.7 bits (104), Expect = 6e-05
Identities = 22/45 (48%), Positives = 30/45 (65%)
Frame = +3
Query: 567 WFYTGDIGRFHPDGCLEIIDRKKDIVKLQHGEYISLGKVEAALAS 611
WF TGD+G HPDG +E+ DR KDI+ + GE IS ++E + S
Sbjct: 24 WFRTGDLGVKHPDGYIELKDRSKDII-ISGGENISSIELEGVIFS 155
>AV772740
Length = 555
Score = 44.3 bits (103), Expect = 7e-05
Identities = 20/52 (38%), Positives = 30/52 (57%)
Frame = -2
Query: 257 KIASFDEVEKLGKESPVEPSLPSKNAVAVVMYTSGSTGLPKGVMITHGNIVA 308
K+ S+ E +G+ P S P + +MYTSG+TG P V++TH NI +
Sbjct: 404 KLYSWKEFLYMGQAKPSTMSPPQAYNICTIMYTSGTTGTPSAVVLTHANITS 249
>TC7805 homologue to PIR|PQ0772|PQ0772 4-coumarate-CoA ligase (clone
GM4CL1B) - soybean (fragment) {Glycine
max;} , partial (41%)
Length = 965
Score = 43.9 bits (102), Expect = 1e-04
Identities = 28/98 (28%), Positives = 51/98 (51%)
Frame = +3
Query: 514 IKLVSWEEGGYLSSDKPMPRGEVVVGGFSVTAGYFKNQDKTDEVFKVDEKGVRWFYTGDI 573
+K++ E G L ++P GE+ + G + GY ++ T +D +G W +TGD+
Sbjct: 102 LKVLDPETGCSLGYNQP---GEICIRGQQIMKGYLNDEKATATT--IDAEG--WLHTGDV 260
Query: 574 GRFHPDGCLEIIDRKKDIVKLQHGEYISLGKVEAALAS 611
G D + I+DR K+++K + G + ++E L S
Sbjct: 261 GYIDDDDEIFIVDRVKELIKFK-GFQVPPAELEGLLVS 371
>BP053582
Length = 516
Score = 41.6 bits (96), Expect = 5e-04
Identities = 20/46 (43%), Positives = 26/46 (56%)
Frame = -1
Query: 547 YFKNQDKTDEVFKVDEKGVRWFYTGDIGRFHPDGCLEIIDRKKDIV 592
Y KN T + FK WF TGD+ HPDG +E+ DR KD++
Sbjct: 465 YLKNLKATQDAFKGG-----WFRTGDLEVKHPDGYIELKDRSKDLI 343
>AV409031
Length = 343
Score = 38.1 bits (87), Expect = 0.005
Identities = 23/77 (29%), Positives = 42/77 (53%)
Frame = +3
Query: 283 VAVVMYTSGSTGLPKGVMITHGNIVATTAAVMTVIPNLGSKDVYLAYLPLAHVFEMAAES 342
+A+ ++TSG+T PKGV ++ N+ ++ + +V L D + LPL HV + A
Sbjct: 120 IALFLHTSGTTSRPKGVPLSQHNLASSVRNIQSVY-RLTESDSTVIVLPLFHVHGLIAGL 296
Query: 343 VMLAAGVAIGYGSPMTL 359
+ ++G G+ +TL
Sbjct: 297 LS-----SLGVGAAVTL 332
>TC15125 weakly similar to UP|Q84P20 (Q84P20) Acyl-activating enzyme 12,
partial (48%)
Length = 1164
Score = 37.4 bits (85), Expect = 0.009
Identities = 23/75 (30%), Positives = 42/75 (55%), Gaps = 1/75 (1%)
Frame = +1
Query: 261 FDEVEKLGKESPVEPSLPSKNAVAVVMYTSGSTGLPKGVMITH-GNIVATTAAVMTVIPN 319
++++ G + V + + + + YTSG+T PKGV+ +H G ++T + V+
Sbjct: 616 YEQMVHRGNPNYVPEEIQEEWSPIALNYTSGTTSEPKGVVYSHRGAYLSTLSLVLGW--E 789
Query: 320 LGSKDVYLAYLPLAH 334
+GS+ VYL LP+ H
Sbjct: 790 MGSEPVYLWTLPMFH 834
>BP059692
Length = 486
Score = 37.4 bits (85), Expect = 0.009
Identities = 48/212 (22%), Positives = 83/212 (38%), Gaps = 2/212 (0%)
Frame = -1
Query: 292 STGLPKGVMITHGNIVATTAAVMTVIPNLGSKDVYLAYLPLAHVFEMAAESV-MLAAGVA 350
+TG KGV+ +H N++A V + + ++ +P+ H++ +A + +L +G
Sbjct: 486 TTGPSKGVISSHRNLIAMVEIVRSRFSKEDER-TFICTVPMFHIYGLAVFATGLLISGST 310
Query: 351 IGYGSPMTLTDTSNKVKKGTKGDVTVLKPTLLTAVPAIIDRIRDGVVKKVEEKGGLAKNL 410
I S + D + + K + T L VP I+
Sbjct: 309 IVVLSKFEMHDMFSTIDK--------FRATFLPLVPPIL--------------------- 217
Query: 411 FQIAYKRRLAAVKGSWLGAWGVEKLVWDTIIFKKIRTVLGGNLRFMLCGGAPLSGD-SQQ 469
+A A+KG + + TVL G GAPLS + ++
Sbjct: 216 --VAMINNADAIKGKY--------------DLSSLHTVLSG--------GAPLSKEVTEG 109
Query: 470 FINICVGAPIGQGYGLTETFAGAAFSEADDYS 501
F++ I QGYGLTE+ A +++ D S
Sbjct: 108 FVDKYPNVTILQGYGLTESSGVGASTDSLDES 13
>AW719487
Length = 498
Score = 37.4 bits (85), Expect = 0.009
Identities = 20/61 (32%), Positives = 36/61 (58%)
Frame = +2
Query: 560 VDEKGVRWFYTGDIGRFHPDGCLEIIDRKKDIVKLQHGEYISLGKVEAALASGDYVDSIM 619
+D++G W +TGDIG D L I+DR K+++K + G ++ ++EA + S + +
Sbjct: 29 IDKEG--WLHTGDIGYIDEDDELFIVDRLKELIKYK-GFQVAPAELEALILSHPKISDVA 199
Query: 620 V 620
V
Sbjct: 200 V 202
>AV409483
Length = 358
Score = 33.5 bits (75), Expect = 0.13
Identities = 21/48 (43%), Positives = 29/48 (59%), Gaps = 1/48 (2%)
Frame = +1
Query: 288 YTSGSTGLPKGVMITHGNIVATTAAVMTVIPN-LGSKDVYLAYLPLAH 334
YTSG+T PKGV+ +H A A+ TV+ N + S VYL +P+ H
Sbjct: 145 YTSGTTSSPKGVIYSHRG--AYLNALATVLLNEMRSMPVYLWCVPMFH 282
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,935,735
Number of Sequences: 28460
Number of extensions: 155601
Number of successful extensions: 671
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 661
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 666
length of query: 755
length of database: 4,897,600
effective HSP length: 97
effective length of query: 658
effective length of database: 2,136,980
effective search space: 1406132840
effective search space used: 1406132840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148718.8


