

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148654.3 - phase: 0 /pseudo
(719 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
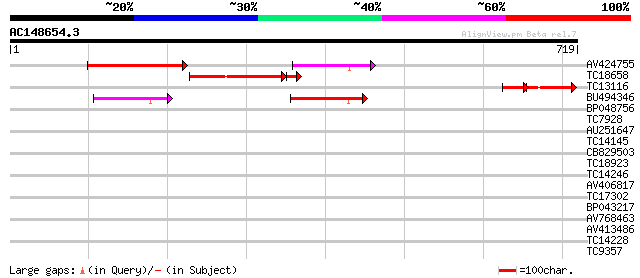
Score E
Sequences producing significant alignments: (bits) Value
AV424755 217 5e-57
TC18658 similar to GB|AAO64894.1|29029030|BT005959 At5g08720 {Ar... 188 3e-53
TC13116 similar to GB|AAO64894.1|29029030|BT005959 At5g08720 {Ar... 64 9e-19
BU494346 70 1e-12
BP048756 37 0.014
TC7928 similar to UP|Q93YR3 (Q93YR3) HSP associated protein like... 30 1.0
AU251647 30 1.3
TC14145 similar to UP|Q7XXS4 (Q7XXS4) Thiamine biosynthetic enzy... 30 1.8
CB829503 30 1.8
TC18923 UP|CAE45594 (CAE45594) S-receptor kinase-like protein 1,... 29 3.0
TC14246 similar to UP|Q84LL6 (Q84LL6) Salt tolerance protein 5, ... 28 3.9
AV406817 28 6.7
TC17302 28 6.7
BP043217 27 8.7
AV768463 27 8.7
AV413486 27 8.7
TC14228 homologue to UP|Q9LLR4 (Q9LLR4) Sec61p, partial (67%) 27 8.7
TC9357 homologue to UP|Q39193 (Q39193) Protein kinase (Protein k... 27 8.7
>AV424755
Length = 383
Score = 217 bits (553), Expect = 5e-57
Identities = 104/127 (81%), Positives = 111/127 (86%)
Frame = +3
Query: 99 LQVVSWRERRVKAEISINADINSVWNALTDYEHLADFIPNLVWSGRIPCPFPGRIWLEQR 158
L VVSWRE+RV+AEISINADI+ +W ALTDY L D IPNLVWSGRI CP PGRIWLEQR
Sbjct: 3 LHVVSWREQRVRAEISINADIHFLWKALTDYYLLVDSIPNLVWSGRIRCPHPGRIWLEQR 182
Query: 159 GFQRAMYWHIEARVVLDLQELLNSEWDRELHFSMVDGDFKKFEGKWSVKSGTRSSSTNLS 218
GFQRAMYWHIEA VVLDLQE++NS WDRELHF MVDGDFKKF+GKWSVKSGT SSST L
Sbjct: 183 GFQRAMYWHIEACVVLDLQEIINSAWDRELHFYMVDGDFKKFDGKWSVKSGTGSSSTTLY 362
Query: 219 YEVNVIP 225
YE NVIP
Sbjct: 363 YEDNVIP 383
Score = 55.8 bits (133), Expect = 2e-08
Identities = 35/111 (31%), Positives = 58/111 (51%), Gaps = 6/111 (5%)
Frame = +3
Query: 359 VVASITVKAPVRDVWNVMSSYETLPEIVPNLAIS-KILSRDNNKVRILQEGCKGLLYMVL 417
V A I++ A + +W ++ Y L + +PNL S +I ++ + Q G + +Y +
Sbjct: 33 VRAEISINADIHFLWKALTDYYLLVDSIPNLVWSGRIRCPHPGRIWLEQRGFQRAMYWHI 212
Query: 418 HARVVLDLCEQL----EQEISFEQAEGDFDSFHGKWTFEQ-LGNHHTLLKY 463
A VVLDL E + ++E+ F +GDF F GKW+ + G+ T L Y
Sbjct: 213 EACVVLDLQEIINSAWDRELHFYMVDGDFKKFDGKWSVKSGTGSSSTTLYY 365
>TC18658 similar to GB|AAO64894.1|29029030|BT005959 At5g08720 {Arabidopsis
thaliana;}, partial (11%)
Length = 524
Score = 188 bits (478), Expect(2) = 3e-53
Identities = 92/122 (75%), Positives = 108/122 (88%)
Frame = +3
Query: 229 FPAIFLERIVRSDLPVNLRALAYRVERNLLGNQKLPQPEDDLHKTSLVVNGSSVKKINGS 288
F AIFLERI+RSDLPVNLRALAYRVERN++GNQ+LP E++LHKT + N S V+KING+
Sbjct: 3 FLAIFLERIIRSDLPVNLRALAYRVERNIIGNQRLPLQENNLHKT-FMPNTSLVEKINGT 179
Query: 289 LCETDKLAPGQDKEGLDTSISGSLPASSSELNSNWGIFGKVCSLDKPCVVDEVHLRRFDG 348
LC +DKL PG++KEGL + ISGSLP SSSE+NSNWG+FGKVC LD+PCVVDEVHLRRFDG
Sbjct: 180 LCGSDKLPPGENKEGLASPISGSLPTSSSEVNSNWGVFGKVCRLDRPCVVDEVHLRRFDG 359
Query: 349 LL 350
LL
Sbjct: 360 LL 365
Score = 38.1 bits (87), Expect(2) = 3e-53
Identities = 18/20 (90%), Positives = 18/20 (90%)
Frame = +2
Query: 351 ENGGVHRCVVASITVKAPVR 370
EN GVH CVVASITVKAPVR
Sbjct: 464 ENRGVHCCVVASITVKAPVR 523
>TC13116 similar to GB|AAO64894.1|29029030|BT005959 At5g08720 {Arabidopsis
thaliana;}, partial (7%)
Length = 519
Score = 63.9 bits (154), Expect(2) = 9e-19
Identities = 38/63 (60%), Positives = 45/63 (71%)
Frame = +3
Query: 656 FERVVSQGKEKKNQTGQQPWQG*EK*SQRISRCR**DQCSI*TLYFSRHTQVAYRTKRVG 715
F V Q +E+ QT QP QG E *S + CR**DQ +I*TLYFSR+TQVAY+T+ VG
Sbjct: 165 FSPSVPQSEEETKQT-TQPCQGQEN*SYSRT*CR**DQGTI*TLYFSRYTQVAYKTETVG 341
Query: 716 HKL 718
HKL
Sbjct: 342 HKL 350
Score = 47.0 bits (110), Expect(2) = 9e-19
Identities = 21/34 (61%), Positives = 25/34 (72%)
Frame = +1
Query: 625 YWDNLENLQDEISRFQRCWGMDPSFMPSRKSFER 658
+ N+ + +ISRFQR WGMDPSFMPS KS ER
Sbjct: 7 FQQNIIHFFMQISRFQRSWGMDPSFMPSSKSLER 108
>BU494346
Length = 575
Score = 70.1 bits (170), Expect = 1e-12
Identities = 41/104 (39%), Positives = 59/104 (56%), Gaps = 4/104 (3%)
Frame = +1
Query: 107 RRVKAEISINADINSVWNALTDYEHLADFIPNLVWSGRIPCPFPGRIWLEQRGFQRAMYW 166
RR+++ ISI+A +++VW+ LTDYE LADFIP L S + L+ A
Sbjct: 37 RRIESRISIDASLDAVWSILTDYERLADFIPGLAVSQLLQKGHNFARLLQIGEQNLAFGI 216
Query: 167 HIEARVVLDLQ----ELLNSEWDRELHFSMVDGDFKKFEGKWSV 206
A+ V+D E L S R++ F M++GDF+ FEGKWS+
Sbjct: 217 KFNAKGVVDCYEKELETLPSGMKRDIEFKMIEGDFQLFEGKWSI 348
Score = 67.0 bits (162), Expect = 1e-11
Identities = 34/106 (32%), Positives = 64/106 (60%), Gaps = 9/106 (8%)
Frame = +1
Query: 357 RCVVASITVKAPVRDVWNVMSSYETLPEIVPNLAISKILSRDNNKVRILQEGCKGLLYMV 416
R + + I++ A + VW++++ YE L + +P LA+S++L + +N R+LQ G + L + +
Sbjct: 37 RRIESRISIDASLDAVWSILTDYERLADFIPGLAVSQLLQKGHNFARLLQIGEQNLAFGI 216
Query: 417 -LHARVVLDLCEQ--------LEQEISFEQAEGDFDSFHGKWTFEQ 453
+A+ V+D E+ ++++I F+ EGDF F GKW+ Q
Sbjct: 217 KFNAKGVVDCYEKELETLPSGMKRDIEFKMIEGDFQLFEGKWSILQ 354
>BP048756
Length = 547
Score = 36.6 bits (83), Expect = 0.014
Identities = 22/69 (31%), Positives = 36/69 (51%), Gaps = 5/69 (7%)
Frame = -3
Query: 513 QNTNSGQQIILSGSGDDNNSSSADDISDCNVQSSSNQRSRVPGLQRD-IEVLK----SEL 567
+N G + G GDD++ SA+D+SD + S++ + +P L D I LK +
Sbjct: 503 KNVYEGVPEKIDGDGDDDDGKSAEDVSDTDSDSANEKYKAIPDLTADQIRGLKFCSSLDA 324
Query: 568 LKFVAEYGQ 576
LKF + Y +
Sbjct: 323 LKFYSSYAK 297
>TC7928 similar to UP|Q93YR3 (Q93YR3) HSP associated protein like, partial
(52%)
Length = 765
Score = 30.4 bits (67), Expect = 1.0
Identities = 20/64 (31%), Positives = 30/64 (46%)
Frame = -1
Query: 488 LPSNLCAIRDYVENQKASQFLEVCEQNTNSGQQIILSGSGDDNNSSSADDISDCNVQSSS 547
LPS + +I + N +AS+ V + G I GS + S + ISD + SSS
Sbjct: 396 LPSEIASIALALANCEASRLSSV---TSTEGSPIFCGGSSSGSTISPSSSISDSTISSSS 226
Query: 548 NQRS 551
+ S
Sbjct: 225 SGSS 214
>AU251647
Length = 390
Score = 30.0 bits (66), Expect = 1.3
Identities = 16/40 (40%), Positives = 24/40 (60%)
Frame = +3
Query: 513 QNTNSGQQIILSGSGDDNNSSSADDISDCNVQSSSNQRSR 552
+N+NSG G D+N S S+++ DC V SS+ QR +
Sbjct: 159 KNSNSGG----GGDHDNNKSFSSEEDEDCEVVSSNLQRDQ 266
>TC14145 similar to UP|Q7XXS4 (Q7XXS4) Thiamine biosynthetic enzyme, partial
(83%)
Length = 1372
Score = 29.6 bits (65), Expect = 1.8
Identities = 17/41 (41%), Positives = 21/41 (50%)
Frame = +2
Query: 8 SSTTTQLRLHTLTLTSMITACRASSSRANPLSLRSSLHSKP 48
S+T Q LT+ +M T SS +NP S SS H KP
Sbjct: 80 SNTLLQNLFTQLTMAAMATTTLTSSLISNPKSSFSSFHGKP 202
>CB829503
Length = 575
Score = 29.6 bits (65), Expect = 1.8
Identities = 15/38 (39%), Positives = 24/38 (62%)
Frame = +1
Query: 534 SADDISDCNVQSSSNQRSRVPGLQRDIEVLKSELLKFV 571
SA SD N+ + ++ RS P L+ + LKSELL+++
Sbjct: 22 SAAPGSDTNIAAMASSRSTCPDLEDEDVDLKSELLRYI 135
>TC18923 UP|CAE45594 (CAE45594) S-receptor kinase-like protein 1, complete
Length = 2061
Score = 28.9 bits (63), Expect = 3.0
Identities = 27/88 (30%), Positives = 36/88 (40%), Gaps = 1/88 (1%)
Frame = -3
Query: 2 PTCHETSSTTTQLRLHTLTLTSMITACRASSSRANPLSLRSSLHSKPSFLSLSLFFPRHF 61
P CH+T S+ + H L + M SS AN S H+ S H
Sbjct: 1162 PECHQTKSSRHYHQYHYLNMHMM-----CKSSFANITS-----HTHQGSCSC------HT 1031
Query: 62 HKSIALSSTTQCKPRSHL-GGNLNNGLE 88
K + L+++ C SHL NLN LE
Sbjct: 1030 RKKMCLATSLLCASESHLHSDNLNFSLE 947
>TC14246 similar to UP|Q84LL6 (Q84LL6) Salt tolerance protein 5, partial
(57%)
Length = 1280
Score = 28.5 bits (62), Expect = 3.9
Identities = 27/79 (34%), Positives = 37/79 (46%), Gaps = 8/79 (10%)
Frame = -1
Query: 9 STTTQLRL-----HTLTLTSMITACRASSSRANPLSLRSSLHSKPSFLSLSL---FFPRH 60
S TT L L TLT+TS + SR+ PL L ++ + PSFLS S FF
Sbjct: 593 SQTTNLDLVPCGTGTLTVTSCRV*VQEYFSRSMPLPLFGTMAAVPSFLSFSSALGFFSAS 414
Query: 61 FHKSIALSSTTQCKPRSHL 79
F+ A + ++ S L
Sbjct: 413 FNLFSAFAFSSDALSSSSL 357
>AV406817
Length = 420
Score = 27.7 bits (60), Expect = 6.7
Identities = 16/40 (40%), Positives = 23/40 (57%), Gaps = 5/40 (12%)
Frame = -3
Query: 25 ITACRASSSRANPL-----SLRSSLHSKPSFLSLSLFFPR 59
I R +S R+N S RSS + PSF++ +L+FPR
Sbjct: 238 IARSRIASDRSNEFPALL*SSRSSASTLPSFVNPTLYFPR 119
>TC17302
Length = 820
Score = 27.7 bits (60), Expect = 6.7
Identities = 26/108 (24%), Positives = 51/108 (47%), Gaps = 4/108 (3%)
Frame = +2
Query: 523 LSGSGDDNNSSSADDISDCNVQSSSNQRSRVPGLQRDIE-VLKSELLK---FVAEYGQEG 578
L+GS + ++ + + SSS QR + + + IE +L LL F+ ++ Q
Sbjct: 56 LAGSVQGDGDAAGNGAESDSRGSSSYQRYDIQQVAKWIEQILPFSLLLLIVFIRQHLQGF 235
Query: 579 FMPMRKQLRLHGRVDIEKAITRMGGFRKIATIMNLSLAYKYRKPKGYW 626
F+ + + +I K T + G RK++ ++ +S A+ + YW
Sbjct: 236 FVTIWISAVMFKSNEIVKKQTALKGDRKVSVLVGISFAFILQVTCIYW 379
>BP043217
Length = 446
Score = 27.3 bits (59), Expect = 8.7
Identities = 16/39 (41%), Positives = 24/39 (61%), Gaps = 3/39 (7%)
Frame = +3
Query: 28 CRASSSRANPLSL---RSSLHSKPSFLSLSLFFPRHFHK 63
C +S S + L L ++SL+++ S SLSL RHFH+
Sbjct: 282 CSSSKSARSQLMLALGQTSLNTEASSGSLSLIAERHFHQ 398
>AV768463
Length = 536
Score = 27.3 bits (59), Expect = 8.7
Identities = 14/48 (29%), Positives = 23/48 (47%)
Frame = +2
Query: 102 VSWRERRVKAEISINADINSVWNALTDYEHLADFIPNLVWSGRIPCPF 149
+SW+++ V +I N +I + NA + L + W RIP F
Sbjct: 71 LSWKDKTVMTQILKNKNIFLI*NASSTCSALQKLYIEITWQQRIPPRF 214
>AV413486
Length = 402
Score = 27.3 bits (59), Expect = 8.7
Identities = 8/28 (28%), Positives = 17/28 (60%)
Frame = -2
Query: 114 SINADINSVWNALTDYEHLADFIPNLVW 141
++ +N VWN + D E+ + +P ++W
Sbjct: 323 ALRRHLNKVWNGILDSENPLNCLPEILW 240
>TC14228 homologue to UP|Q9LLR4 (Q9LLR4) Sec61p, partial (67%)
Length = 1268
Score = 27.3 bits (59), Expect = 8.7
Identities = 15/59 (25%), Positives = 28/59 (47%)
Frame = +2
Query: 272 KTSLVVNGSSVKKINGSLCETDKLAPGQDKEGLDTSISGSLPASSSELNSNWGIFGKVC 330
KT + V+GSS + + L E + PG + L ++ +P +++ FG +C
Sbjct: 749 KTWIEVSGSSARDVAKQLKEQQMVMPGHRESNLQKELNRYIPTAAA--------FGGIC 901
>TC9357 homologue to UP|Q39193 (Q39193) Protein kinase (Protein kinase, 41K)
, partial (71%)
Length = 1181
Score = 27.3 bits (59), Expect = 8.7
Identities = 9/27 (33%), Positives = 17/27 (62%)
Frame = -1
Query: 119 INSVWNALTDYEHLADFIPNLVWSGRI 145
+N VWN + D ++ + +P ++W RI
Sbjct: 1037 LNKVWNGILDTQNSLNCLPEILWFIRI 957
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.137 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,892,898
Number of Sequences: 28460
Number of extensions: 188104
Number of successful extensions: 1049
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 1035
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1046
length of query: 719
length of database: 4,897,600
effective HSP length: 97
effective length of query: 622
effective length of database: 2,136,980
effective search space: 1329201560
effective search space used: 1329201560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148654.3


