

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148651.11 - phase: 0
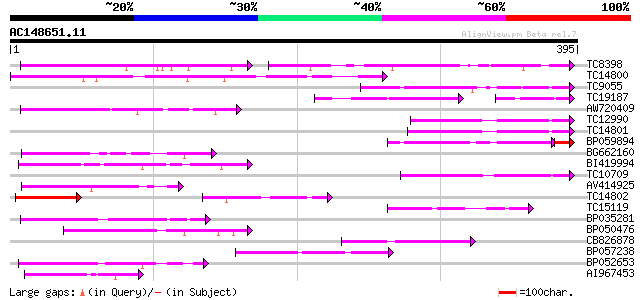
(395 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8398 weakly similar to UP|Q84KK1 (Q84KK1) S haplotype-specific... 91 4e-34
TC14800 weakly similar to UP|Q9FHP3 (Q9FHP3) Genomic DNA, chromo... 113 6e-26
TC9055 92 1e-19
TC19187 80 6e-18
AW720409 84 4e-17
TC12990 79 2e-15
TC14801 78 3e-15
BP059894 70 4e-14
BG662160 74 6e-14
BI419994 71 3e-13
TC10709 64 3e-11
AV414925 64 6e-11
TC14802 weakly similar to UP|CAD56661 (CAD56661) S locus F-box (... 61 4e-10
TC15119 58 2e-09
BP035281 57 7e-09
BP050476 57 7e-09
CB826878 56 9e-09
BP057238 54 3e-08
BP052653 54 5e-08
AI967453 54 6e-08
>TC8398 weakly similar to UP|Q84KK1 (Q84KK1) S haplotype-specific F-box
protein a, partial (9%)
Length = 1602
Score = 90.5 bits (223), Expect(2) = 4e-34
Identities = 76/224 (33%), Positives = 104/224 (45%), Gaps = 11/224 (4%)
Frame = +1
Query: 181 RSVVKVFSMRDNCWRNIQCFPVLPLYMF----VSTQNGVYFSSTVNWLALQDYFGLDYFH 236
+SV V++M D CWR IQ FP + QNGVY + T+NW+
Sbjct: 724 QSVANVYNMGDGCWRRIQSFPDFTREHWGRGTAPDQNGVYLNGTINWV----------LR 873
Query: 237 LNYSSITPEKYVILSLDLSTETYTQLLLP----RGFNKVSRHQPKLAVLMDCLCFGHDYE 292
LN + Y I+SLDL +E ++L LP G V P L VL D LC H+
Sbjct: 874 LNIA------YDIISLDLGSEVCSRLSLPCFPPPGPKCVYDLPPILGVLKDSLCVLHNNN 1035
Query: 293 ETYFVIWQMKDFGVQSSWIQLFKITYQNFFSYYCDFARESKWLDFLPLCLSKNGDTLILA 352
+ FVIW+M +FG SW LF NF DF +E + F S+N + L++
Sbjct: 1036DKTFVIWKMNEFGAHESWTPLF-----NF-----DFKKEDLY-RFDKFFFSENDEVLLMT 1182
Query: 353 NNQE---NEAFIYNRRDDRVEKIGITNKNMLWFEAWDYVESLVS 393
++ N + I + +DRV N W +YVESLVS
Sbjct: 1183LTKQVLYNHSDITFKFNDRVH-----NYIHDWNNIENYVESLVS 1299
Score = 71.2 bits (173), Expect(2) = 4e-34
Identities = 56/203 (27%), Positives = 94/203 (45%), Gaps = 41/203 (20%)
Frame = +3
Query: 8 VLPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQHN 67
+L EL+ +ILSLLPVK L+RFRCV K + IS+P F+++HL+ S+ Q
Sbjct: 75 ILIDELIWEILSLLPVKSLVRFRCVCKSWKLTISNPQFMKLHLRRSSAKAAADFADSQVV 254
Query: 68 FNSFDENVLNLPI----SLLLENSLSTVPYDPYYRLKN--ENPH-----CPWLFA----- 111
+ +++ + P+ ++ + + + P + L+ PH P LF+
Sbjct: 255 VMTKRDHIAHSPLLSKFHIVHDETFTADEPWPVHALREPFNQPHVSICYAPSLFSSQNND 434
Query: 112 ---------GSCNGLICLCLD----IDTSHGSRLCLWNPATRTKSEFDLASQECF----- 153
G+CNGL+ + + +++ C NPAT+ +S+ C+
Sbjct: 435 KLKGHCRLIGACNGLVSVINEGYNSAESATKLSCCNLNPATKLRSQVSPTLSLCYKQFHS 614
Query: 154 -------VFAFGYDNLNGNYKVI 169
+F FGYD LN YKV+
Sbjct: 615 LIHSVFRIFGFGYDPLNDTYKVV 683
>TC14800 weakly similar to UP|Q9FHP3 (Q9FHP3) Genomic DNA, chromosome 5, TAC
clone:K14B20, partial (7%)
Length = 782
Score = 113 bits (282), Expect = 6e-26
Identities = 88/274 (32%), Positives = 134/274 (48%), Gaps = 11/274 (4%)
Frame = +3
Query: 1 MEDRHQSVLPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHL--KNSARNPN 58
ME H LP EL+ +ILS LPVK L++FR V+K + + ISDP F+++HL + S RN +
Sbjct: 24 MEPLHLLCLPDELIIEILSWLPVKSLLQFRVVSKTWKSFISDPQFVKLHLLHRLSFRNAD 203
Query: 59 L---MVIARQHNFNSFDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSCN 115
++ + H + + + +S LLE+ P + + + + F G+CN
Sbjct: 204 FEHTSLLIKCHTDDFGRPYISSRTVSSLLES--------PSAIVASRSCISGYDFIGTCN 359
Query: 116 GLICLCL----DIDTSHGSRLCLWNPATRTKSEFDLA--SQECFVFAFGYDNLNGNYKVI 169
GL+ L + +T++ S++ WNPATRT S+ S FGYD + YKV+
Sbjct: 360 GLVSLRKLNYDESNTNNFSQVRFWNPATRTMSQDSPPSWSPRNLHLGFGYDCSSDTYKVV 539
Query: 170 AFDIKVKSGNARSVVKVFSMRDNCWRNIQCFPVLPLYMFVSTQNGVYFSSTVNWLALQDY 229
++V V++M DNCWR IQ P P+++ + VY S+T+NWLA
Sbjct: 540 GMI------PGLTMVNVYNMGDNCWRTIQISPHAPMHL---QGSAVYVSNTLNWLA---- 680
Query: 230 FGLDYFHLNYSSITPEKYVILSLDLSTETYTQLL 263
Y I+S DL E Q L
Sbjct: 681 -------------GTNPYFIVSFDLEKEECAQQL 743
>TC9055
Length = 577
Score = 92.4 bits (228), Expect = 1e-19
Identities = 55/152 (36%), Positives = 89/152 (58%), Gaps = 3/152 (1%)
Frame = +2
Query: 245 EKYVILSLDLSTETYTQLLLPRGFNKVSRHQPKLAVLMDCLCFGHDYEETYFVIWQMKDF 304
E+ VI+SLD+ E Y L LP+G +++ ++ VL +CLC +D++ T+FV W+M ++
Sbjct: 41 EQLVIVSLDMRQEAYRLLSLPQGTSELP--SAEIQVLGNCLCLFYDFKRTHFVAWKMSEY 214
Query: 305 GVQSSWIQLFKITYQNF---FSYYCDFARESKWLDFLPLCLSKNGDTLILANNQENEAFI 361
G+ SW L I+Y++ +Y ++WL +CL +G +LA Q+ I
Sbjct: 215 GLPESWTPLLTISYEHLRWDHRFYL-----NQWL----ICLCGDGHIFMLAKKQK-LVII 364
Query: 362 YNRRDDRVEKIGITNKNMLWFEAWDYVESLVS 393
YN D+ V+ + + + N LW +A DYVESLVS
Sbjct: 365 YNLSDNSVKYVELAS-NKLWLDANDYVESLVS 457
>TC19187
Length = 632
Score = 80.5 bits (197), Expect(2) = 6e-18
Identities = 41/104 (39%), Positives = 59/104 (56%)
Frame = +1
Query: 213 NGVYFSSTVNWLALQDYFGLDYFHLNYSSITPEKYVILSLDLSTETYTQLLLPRGFNKVS 272
+G + S T+NW A NYS ++I+SLDL ETY ++L P + K
Sbjct: 4 SGKFVSGTLNWAA------------NYSVGVSSLWIIVSLDLQKETYKEIL-PPDYEKEE 144
Query: 273 RHQPKLAVLMDCLCFGHDYEETYFVIWQMKDFGVQSSWIQLFKI 316
P L+VL CLC +D++ T FV+W MKD+GV+ SW++L I
Sbjct: 145 CSTPTLSVLKGCLCMNYDHKRTDFVVWMMKDYGVRESWVKLVTI 276
Score = 26.6 bits (57), Expect(2) = 6e-18
Identities = 16/55 (29%), Positives = 30/55 (54%)
Frame = +2
Query: 339 PLCLSKNGDTLILANNQENEAFIYNRRDDRVEKIGITNKNMLWFEAWDYVESLVS 393
P +S+NG+ L++ E + +Y+ RD + + WF+A Y+E+L+S
Sbjct: 317 PYFISENGEVLLMF---EFDLVMYDPRDQSFRYPRLESGKG-WFDAEVYIETLIS 469
>AW720409
Length = 575
Score = 84.0 bits (206), Expect = 4e-17
Identities = 61/166 (36%), Positives = 83/166 (49%), Gaps = 12/166 (7%)
Frame = +3
Query: 8 VLPSELMTKILSLLPVKPLMRFRCVNKFYNTLI-SDPHFIQMHLKNSARNPNLMVIARQH 66
VLP EL +ILS LPVK LMRF CV+K + +LI D F ++HL S +N N +++ Q
Sbjct: 90 VLPFELWIEILSWLPVKTLMRFSCVSKSWKSLIYQDRDFKKLHLDRSPKNNNQVILTLQK 269
Query: 67 NFNSFDENVLNLPISLLLENS-------LSTVPYDPYYRLKNENPHCPWLFAGSCNGLIC 119
S P+ LL++ + +P++ + +N H GSCNGL+C
Sbjct: 270 PLFS-GRTFFPFPVRRLLQDQEPSSSSIIDDIPFEEEDEDEEDNYHDT---IGSCNGLVC 437
Query: 120 LCLDIDTSHGSRLCLWNPATRTK----SEFDLASQECFVFAFGYDN 161
L D HG LWNPATR + D FAFGYD+
Sbjct: 438 LYGIND--HGLWFRLWNPATRFRFHKSPPLDAVIGSTLHFAFGYDH 569
>TC12990
Length = 518
Score = 78.6 bits (192), Expect = 2e-15
Identities = 45/115 (39%), Positives = 66/115 (57%), Gaps = 1/115 (0%)
Frame = +1
Query: 280 VLMDCLCFGHDYEETY-FVIWQMKDFGVQSSWIQLFKITYQNFFSYYCDFARESKWLDFL 338
VL CLC + +ET+ FV+WQMK+FGV SW QLF I + FA
Sbjct: 4 VLGGCLCISQNNKETHSFVVWQMKEFGVHESWTQLFNIIVHERILHTPCFA--------- 156
Query: 339 PLCLSKNGDTLILANNQENEAFIYNRRDDRVEKIGITNKNMLWFEAWDYVESLVS 393
+C+S+NGD L+ A + ++A +YN+R+ + E I N N++ +Y+ESLVS
Sbjct: 157 -MCMSENGDALLFAKSGVSQAVLYNQREKKFEDTKIGN-NIVGSYVRNYIESLVS 315
>TC14801
Length = 595
Score = 77.8 bits (190), Expect = 3e-15
Identities = 44/117 (37%), Positives = 68/117 (57%), Gaps = 1/117 (0%)
Frame = +2
Query: 278 LAVLMDCLCFGHDYEETY-FVIWQMKDFGVQSSWIQLFKITYQNFFSYYCDFARESKWLD 336
L VL CLC ++ET+ FV+ QMK++GV SW QLF I + FA
Sbjct: 101 LGVLRGCLCISQYHKETHSFVVSQMKEYGVNESWTQLFNIIVHERILHSPCFA------- 259
Query: 337 FLPLCLSKNGDTLILANNQENEAFIYNRRDDRVEKIGITNKNMLWFEAWDYVESLVS 393
+C+S+NGD L+ A + ++A +YN+RD+++E I N N++ +Y+ESL+S
Sbjct: 260 ---MCMSENGDVLLFAESGVSQAVLYNQRDNKLEDTKIAN-NIVGSYVRNYIESLIS 418
>BP059894
Length = 474
Score = 70.1 bits (170), Expect(2) = 4e-14
Identities = 40/119 (33%), Positives = 66/119 (54%), Gaps = 1/119 (0%)
Frame = -3
Query: 264 LPRGFNKVSRHQPKLAVLMDCLCFGHDYEETYFVIWQMKDFGVQSSWIQLFKITYQNFFS 323
LP+G +++ ++ VL +CLC HD T+FV W+M ++GV SW ++ I+YQ+F
Sbjct: 469 LPKGTSELP--YVEIQVLGNCLCLFHDDNRTHFVGWKMTEYGVPESWTRMLSISYQHF-- 302
Query: 324 YYC-DFARESKWLDFLPLCLSKNGDTLILANNQENEAFIYNRRDDRVEKIGITNKNMLW 381
C D +WL +CL +G+ L+LA ++ I+N D+ V+ I K+ W
Sbjct: 301 -QCNDVILRHRWL----VCLCGDGNILMLAKDEGGLVIIFNLSDNSVKYIKFPPKDFGW 140
Score = 24.3 bits (51), Expect(2) = 4e-14
Identities = 10/14 (71%), Positives = 11/14 (78%)
Frame = -2
Query: 380 LWFEAWDYVESLVS 393
LW A DYVESL+S
Sbjct: 149 LWLGAEDYVESLIS 108
>BG662160
Length = 445
Score = 73.6 bits (179), Expect = 6e-14
Identities = 50/139 (35%), Positives = 73/139 (51%), Gaps = 3/139 (2%)
Frame = +2
Query: 9 LPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQHNF 68
LP EL +ILS LPVK L+RFRCV+K + ++ISD FI++HL S+ R +F
Sbjct: 23 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSST------TRNTDF 184
Query: 69 NSFDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLICL---CLDID 125
++ ++++ P S ST+ Y F G+CNGL+CL D
Sbjct: 185 -AYLQSLITSPRK---SRSASTIAIPDDYD-----------FCGTCNGLVCLHSSKYDRK 319
Query: 126 TSHGSRLCLWNPATRTKSE 144
+ S + LWNPA R+ S+
Sbjct: 320 DKYTSHVRLWNPAMRSMSQ 376
>BI419994
Length = 549
Score = 71.2 bits (173), Expect = 3e-13
Identities = 58/174 (33%), Positives = 84/174 (47%), Gaps = 11/174 (6%)
Frame = +2
Query: 7 SVLPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQH 66
S LP EL+ +IL LPV+ L+RF+ V K + LISDP F + H N A +P + +
Sbjct: 41 SHLPDELILQILLRLPVRSLLRFKTVCKSWRYLISDPQFTKSHF-NLAASPTHRIFL--N 211
Query: 67 NFNSFDENVLNLPISLLLENSLSTV------PYDPYYRLKNENPHCPWLFAGSCNGLICL 120
N N F L+ SL E+SL + P+ +R + L GSC G + L
Sbjct: 212 NTNGF*IESLDTDSSLHDESSLVHLKFPLAPPHPDNHRFGSSVTGLKVL--GSCRGFVLL 385
Query: 121 CLDIDTSHGSRLCLWNPATRTKSEFD-----LASQECFVFAFGYDNLNGNYKVI 169
+H + +WNP+T + LASQ F++ GYD N +Y V+
Sbjct: 386 -----ANHQGNVVVWNPSTGVRRRVPEWCSYLASQCEFLYGIGYDESNDDYLVV 532
>TC10709
Length = 578
Score = 64.3 bits (155), Expect = 3e-11
Identities = 38/121 (31%), Positives = 61/121 (50%)
Frame = +3
Query: 273 RHQPKLAVLMDCLCFGHDYEETYFVIWQMKDFGVQSSWIQLFKITYQNFFSYYCDFARES 332
R +P L VL + + HD+ + +W+M +G SW QL I+Y+ +
Sbjct: 3 RFEPSLKVLGNHMYLFHDHNSIHLFVWKMVKYGALESWTQLLSISYERLHCIGFPYIP-- 176
Query: 333 KWLDFLPLCLSKNGDTLILANNQENEAFIYNRRDDRVEKIGITNKNMLWFEAWDYVESLV 392
+P+ L ++GD L++A + E YN RD +E I N+ + W +A+ YV SLV
Sbjct: 177 -----VPMFLLEDGDILMIAITENLEFIKYNIRDYSMEYIPNPNE-VFWMDAYGYVPSLV 338
Query: 393 S 393
S
Sbjct: 339 S 341
>AV414925
Length = 414
Score = 63.5 bits (153), Expect = 6e-11
Identities = 38/115 (33%), Positives = 62/115 (53%), Gaps = 2/115 (1%)
Frame = +1
Query: 9 LPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSAR--NPNLMVIARQH 66
+P E++ ILS LPV L+RFR ++K + +LI HF+++HL NS NPNL + +H
Sbjct: 97 IPVEVIADILSRLPVTSLLRFRSISKSWRSLIDSKHFMKLHLNNSLTNPNPNLTNLILRH 276
Query: 67 NFNSFDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLICLC 121
N + + + ++ ++ L + L Y N GSC+GL+C+C
Sbjct: 277 NTDLYRADFPSIGAAVNLNHPLMC-----YSNRIN--------ILGSCHGLLCIC 402
>TC14802 weakly similar to UP|CAD56661 (CAD56661) S locus F-box (SLF)-S4
protein, partial (10%)
Length = 551
Score = 60.8 bits (146), Expect = 4e-10
Identities = 36/93 (38%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Frame = +3
Query: 135 WNPATRTKSEFDLAS--QECFVFAFGYDNLNGNYKVIAFDIKVKSGNARSVVKVFSMRDN 192
WNPATRT S+ S FGYD + YKV+ + ++V V++M DN
Sbjct: 258 WNPATRTMSQDSPPSWSPRNLHLGFGYDCSSDTYKVVGMIPGL------TMVNVYNMGDN 419
Query: 193 CWRNIQCFPVLPLYMFVSTQNGVYFSSTVNWLA 225
CWR IQ P P+++ + VY S+T+NWLA
Sbjct: 420 CWRTIQISPHAPMHL---QGSAVYVSNTLNWLA 509
Score = 53.1 bits (126), Expect = 8e-08
Identities = 24/46 (52%), Positives = 34/46 (73%)
Frame = +3
Query: 5 HQSVLPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHL 50
H LP EL+ +ILS LPVK L++FR V+K + + ISDP F+++HL
Sbjct: 9 HLLCLPDELIIEILSWLPVKSLLQFRVVSKTWKSFISDPQFVKLHL 146
>TC15119
Length = 520
Score = 58.2 bits (139), Expect = 2e-09
Identities = 34/102 (33%), Positives = 53/102 (51%)
Frame = +3
Query: 264 LPRGFNKVSRHQPKLAVLMDCLCFGHDYEETYFVIWQMKDFGVQSSWIQLFKITYQNFFS 323
LP ++ +P L +L +CLC H++ T +WQM+ FGV+ SW QL KIT
Sbjct: 6 LPSNYDYEKHGEPTLVILKNCLCACHEFLVT---VWQMEKFGVRESWTQLLKIT------ 158
Query: 324 YYCDFARESKWLDFLPLCLSKNGDTLILANNQENEAFIYNRR 365
+ L+ +PLC+S+N ++L + E F +N R
Sbjct: 159 --------NIHLNAVPLCMSEN--DVVLLSKSERIFFHWNHR 254
>BP035281
Length = 498
Score = 56.6 bits (135), Expect = 7e-09
Identities = 46/134 (34%), Positives = 71/134 (52%), Gaps = 1/134 (0%)
Frame = +2
Query: 8 VLPSELMTKILSLLPVKPLMRFRCVNKFYNTLIS-DPHFIQMHLKNSARNPNLMVIARQH 66
V PSEL+ +ILS LPV L+RF+CV K + +LIS + F ++ L+ ++ N +++
Sbjct: 119 VFPSELVIEILSWLPVLSLIRFKCVCKSWKSLISHNKAFAKLQLERTSLKINHVIL---- 286
Query: 67 NFNSFDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLICLCLDIDT 126
S D + + + LLE+ S + D L+ + + GSCNGLICL D
Sbjct: 287 --TSSDSSFIPCSLPRLLEDPSSIMDQDQDRCLRLDKFEYNEVI-GSCNGLICLHADY-C 454
Query: 127 SHGSRLCLWNPATR 140
+ G NP+TR
Sbjct: 455 APGFCFRPCNPSTR 496
>BP050476
Length = 520
Score = 56.6 bits (135), Expect = 7e-09
Identities = 46/147 (31%), Positives = 72/147 (48%), Gaps = 15/147 (10%)
Frame = +2
Query: 38 TLISDPHFIQMHLKNSARNPNLMV-IARQ-HNFNSFDENVLNLPISLLLENSLSTVPYDP 95
T D F+++HL S +N ++++ IA ++F + D V+ + L+E+ S +
Sbjct: 5 TYHDDSSFVKLHLNRSPKNTHILLNIANDPYDFENDDTWVVPSSVCCLIEDLSSMIDAKG 184
Query: 96 YYRLKNENPHCPWLFAGSCNGLICL--CLDIDTSHGSRLCLWNPATRTKSE----FDLAS 149
Y LK+ + L GS NGLIC D+ + LWNPAT KS+ F+L+
Sbjct: 185 CYLLKDGH-----LVIGSSNGLICFGNFYDVGPIEEFWVQLWNPATHLKSKKSPTFNLSM 349
Query: 150 QECFV-------FAFGYDNLNGNYKVI 169
+ FGYDNL+ YKV+
Sbjct: 350 RTSVDAPPGKVNLGFGYDNLHDTYKVV 430
>CB826878
Length = 635
Score = 56.2 bits (134), Expect = 9e-09
Identities = 37/95 (38%), Positives = 48/95 (49%), Gaps = 2/95 (2%)
Frame = +3
Query: 232 LDYFHLNYSSITPEKYVILSLDLSTETYTQLLLPRGFNKVSRHQPKLAVLMDCLCFGHDY 291
LD+F+ NY+ I T+ Q L N + P L VL D LC +
Sbjct: 267 LDFFYANYNLIMQLGNTARCSVSGTQLPEQCL-----NSPHLYMPILGVLRDSLCVSQND 431
Query: 292 EETY-FVIWQMKDFGVQSSWIQLFKIT-YQNFFSY 324
++T FV+WQMK+FGV SW QLF I NFFS+
Sbjct: 432 KKTRNFVVWQMKEFGVHKSWTQLFNIIGDDNFFSW 536
Score = 29.3 bits (64), Expect = 1.2
Identities = 11/16 (68%), Positives = 12/16 (74%)
Frame = +2
Query: 130 SRLCLWNPATRTKSEF 145
S LC WNPATRT +F
Sbjct: 320 SLLCFWNPATRTMPQF 367
Score = 26.6 bits (57), Expect = 7.7
Identities = 13/33 (39%), Positives = 21/33 (63%), Gaps = 1/33 (3%)
Frame = +3
Query: 31 CVNKFYNTLISDPHFIQMHLKN-SARNPNLMVI 62
C N+ +LISD F+++HL S+RN + +I
Sbjct: 105 CQNQSCKSLISDSAFVKLHLHR*SSRNADFSMI 203
>BP057238
Length = 434
Score = 54.3 bits (129), Expect = 3e-08
Identities = 33/110 (30%), Positives = 49/110 (44%)
Frame = -2
Query: 158 GYDNLNGNYKVIAFDIKVKSGNARSVVKVFSMRDNCWRNIQCFPVLPLYMFVSTQNGVYF 217
GYD YK + + V+ M D+CWR I P + + +G +
Sbjct: 433 GYDESRDTYKAVMALCHSIEPEPKMETTVYCMGDSCWRKISSSP--SSSVLLQQFDGQFV 260
Query: 218 SSTVNWLALQDYFGLDYFHLNYSSITPEKYVILSLDLSTETYTQLLLPRG 267
VNWLAL + G +Y + S+T ++ VI+ + E YT L LP G
Sbjct: 259 GGCVNWLALDNLNGPNY---EWQSVTLDQLVIVCFHMREEAYTYLSLPEG 119
>BP052653
Length = 552
Score = 53.9 bits (128), Expect = 5e-08
Identities = 41/134 (30%), Positives = 65/134 (47%), Gaps = 2/134 (1%)
Frame = +1
Query: 7 SVLPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQH 66
S+LP +L+ +IL LPV+ L+RF+ V K + +LISDP F + H +A + ++
Sbjct: 181 SILPHDLIVQILLRLPVRSLLRFKSVCKSWLSLISDPQFGKSHFDLAAAPTHRCLV---- 348
Query: 67 NFNSFDENVLNLPISLLLENSLSTV--PYDPYYRLKNENPHCPWLFAGSCNGLICLCLDI 124
N+F L++ S+L ++S + PY Y + GSC G I L
Sbjct: 349 KINNFKLESLDVDASVLDQSSEVNLKFPYTGYVIFHIQ-------ILGSCRGFIALV--- 498
Query: 125 DTSHGSRLCLWNPA 138
G +WNP+
Sbjct: 499 ---RGGDAIVWNPS 531
>AI967453
Length = 456
Score = 53.5 bits (127), Expect = 6e-08
Identities = 36/86 (41%), Positives = 51/86 (58%), Gaps = 3/86 (3%)
Frame = +1
Query: 11 SELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQHNFNS 70
+ L+T+IL LPVK L+RF+ V KF+ +LISDPHF H + +A P L+ I H +
Sbjct: 10 TSLITEILLRLPVKSLVRFKAVCKFWRSLISDPHFATSHFERAA--PRLL-IRTDHGIRT 180
Query: 71 FD--ENVLNLPISLLLENSLS-TVPY 93
D E++ IS L++ TVPY
Sbjct: 181MDLEESLHPDRISELIDYDFPYTVPY 258
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.325 0.139 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,793,131
Number of Sequences: 28460
Number of extensions: 152341
Number of successful extensions: 1058
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 1027
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1033
length of query: 395
length of database: 4,897,600
effective HSP length: 92
effective length of query: 303
effective length of database: 2,279,280
effective search space: 690621840
effective search space used: 690621840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC148651.11


