

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148607.16 - phase: 0 /pseudo
(619 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
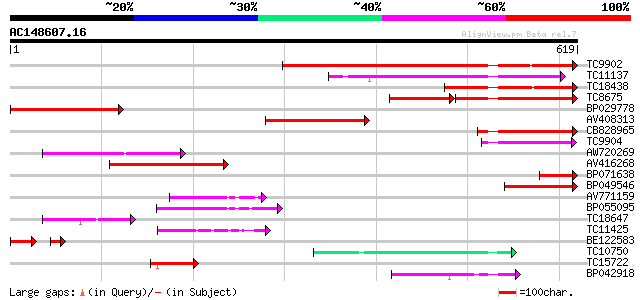
Score E
Sequences producing significant alignments: (bits) Value
TC9902 similar to UP|BAD07483 (BAD07483) PDR-type ABC transporte... 437 e-123
TC11137 similar to GB|CAD59563.1|27368811|OSA535041 PDR-like ABC... 189 1e-48
TC18438 similar to UP|Q8GU88 (Q8GU88) PDR-like ABC transporter, ... 186 1e-47
TC8675 weakly similar to UP|Q9LFH0 (Q9LFH0) PDR9 ABC transporter... 128 2e-47
BP029778 177 6e-45
AV408313 167 6e-42
CB828965 131 4e-31
TC9904 weakly similar to UP|Q8GU88 (Q8GU88) PDR-like ABC transpo... 99 2e-21
AW720269 87 6e-18
AV416268 78 4e-15
BP071638 74 5e-14
BP049546 72 3e-13
AV771159 70 1e-12
BP055095 59 2e-09
TC18647 similar to UP|Q9ASR9 (Q9ASR9) At2g01320/F10A8.20, partia... 58 5e-09
TC11425 similar to UP|Q9M1Q9 (Q9M1Q9) P-glycoprotein-like proeti... 46 2e-05
BE122583 40 6e-05
TC10750 similar to UP|AAP80385 (AAP80385) ABC transporter, parti... 44 6e-05
TC15722 similar to UP|Q9FMU0 (Q9FMU0) Similarity to ABC transpor... 43 2e-04
BP042918 42 3e-04
>TC9902 similar to UP|BAD07483 (BAD07483) PDR-type ABC transporter 1,
partial (19%)
Length = 1207
Score = 437 bits (1123), Expect = e-123
Identities = 197/322 (61%), Positives = 255/322 (79%)
Frame = +1
Query: 298 KNSELYRRNKALIKELSAPAPCSKDLYFPSRYSRSFFTQCIACLWKQHWSYWRNPEYNAI 357
KNS+L+RRNK LI+EL PAP SKDLYF +++S+ F QC ACLWKQ WSYWRNP Y A+
Sbjct: 1 KNSDLFRRNKQLIQELGEPAPDSKDLYFATQFSQPFLIQCQACLWKQRWSYWRNPPYTAV 180
Query: 358 RFLYSTAVAVLLGSMFWNLGSKIEKDQDLFNAMGSMYAAVILIGAMNSNSVQPVVGVERT 417
RF ++T +AV+ G++FW+LG K ++ QDL NA+GSMY+AV+ +G NS+SVQPVV VERT
Sbjct: 181 RFFFTTFIAVMFGTIFWDLGGKHKRRQDLLNAVGSMYSAVLFLGVQNSSSVQPVVSVERT 360
Query: 418 VFYRERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGFEWTLVKVVWCLFFMY 477
VFYRE+AA MYSA PYA AQ++++LPY+F QAV YG++VY MIGF+WT K W LFFMY
Sbjct: 361 VFYREKAAGMYSALPYAFAQILVELPYIFFQAVTYGVIVYAMIGFDWTAEKFFWYLFFMY 540
Query: 478 FTFLYFTFYGMMSVAMTPNNHISIIVSSAFYSVWNLFSGFVLPRPGK**LISVILSIPVW 537
FT LYFTFYGMM VA+TPN+H++ IV++AFY++WNLFSGFV+PRP SIPVW
Sbjct: 541 FTLLYFTFYGMMGVAVTPNHHVASIVAAAFYAIWNLFSGFVVPRP----------SIPVW 690
Query: 538 WRWYSWANPVAWSLYGLVTSQYGDVKQNIETSDGRQTVEDFLRNYFGFKHDFLGVVALVN 597
WRWY WA PVAW++YGL+ SQ+GD+ ++T +G +TV+ FL +Y+G KH F+GV A+V
Sbjct: 691 WRWYYWACPVAWTIYGLIASQFGDITTVMDT-EGGKTVKMFLEDYYGIKHSFIGVCAVVV 867
Query: 598 IAFPIVFALVFAIAIKMFNFQR 619
I+FA +FA+AIK FNFQ+
Sbjct: 868 PGVAILFAFIFAVAIKAFNFQK 933
>TC11137 similar to GB|CAD59563.1|27368811|OSA535041 PDR-like ABC
transporter {Oryza sativa (japonica cultivar-group);},
partial (15%)
Length = 786
Score = 189 bits (480), Expect = 1e-48
Identities = 95/264 (35%), Positives = 144/264 (53%), Gaps = 6/264 (2%)
Frame = +1
Query: 349 WRNPEYNAIRFLYSTAVAVLLGSMFWNLGSKIEKDQDLFNAMG----SMYAAVILIGAMN 404
WR+P+YN +RF + + L S K+ K + + L+
Sbjct: 16 WRSPDYNLVRFSFPCS----LPSWLVLFSGKLAKTRRALPT*AWS*EHCMLLLFLLELTT 183
Query: 405 SNSVQPVVGVERTVFYRERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGFEW 464
+ + ++ + MY+ PYA+AQV ++PYVF Q Y ++VY M+ F+W
Sbjct: 184 AKLFNQLWPLKELYSIEKGLLAMYAPLPYAIAQVFTEIPYVFAQTTFYSLIVYAMVSFDW 363
Query: 465 TLVKVVWCLFFMYFTFLYFTFYGMMSVAMTPNNHISIIVSSAFYSVWNLFSGFVLPRPGK 524
+ K W F +F+FLYFT+YGMM+V++TPN+ ++ I ++AFY ++NLFSGF +PRP
Sbjct: 364 KVDKFFWYFFVSFFSFLYFTYYGMMTVSITPNHQVASIFAAAFYGLFNLFSGFFIPRP-- 537
Query: 525 **LISVILSIPVWWRWYSWANPVAWSLYGLVTSQYGDVKQNIETSDGRQ--TVEDFLRNY 582
IP WW WY W PVAW++YGL+ SQY D+ I + Q TV+ ++ +Y
Sbjct: 538 --------KIPGWWVWYYWICPVAWTVYGLIVSQYRDITSPISVAGSTQNFTVKGYIEDY 693
Query: 583 FGFKHDFLGVVALVNIAFPIVFAL 606
+GFK DF+G VA V + F L
Sbjct: 694 YGFKPDFMGPVAAVLVLFTCFLCL 765
>TC18438 similar to UP|Q8GU88 (Q8GU88) PDR-like ABC transporter, partial
(8%)
Length = 522
Score = 186 bits (471), Expect = 1e-47
Identities = 80/145 (55%), Positives = 113/145 (77%)
Frame = +1
Query: 475 FMYFTFLYFTFYGMMSVAMTPNNHISIIVSSAFYSVWNLFSGFVLPRPGK**LISVILSI 534
FM+FT YFT+YGMM+VA+TPN+H++ IV++AFY++WNLFSGFV+PRP I
Sbjct: 1 FMFFTLCYFTYYGMMTVAVTPNHHVASIVAAAFYAIWNLFSGFVVPRP----------RI 150
Query: 535 PVWWRWYSWANPVAWSLYGLVTSQYGDVKQNIETSDGRQTVEDFLRNYFGFKHDFLGVVA 594
PVWWRWY WA PVAW++YG+V SQ+GD++ +E+ D +V++F+R+YFG KHDF+GV A
Sbjct: 151 PVWWRWYYWACPVAWTIYGMVASQFGDIEHILESDD--VSVKEFIRSYFGMKHDFIGVCA 324
Query: 595 LVNIAFPIVFALVFAIAIKMFNFQR 619
+V + F + FA +FA++ K+FNF R
Sbjct: 325 VVVVGFAVGFAFIFAVSFKVFNFPR 399
>TC8675 weakly similar to UP|Q9LFH0 (Q9LFH0) PDR9 ABC transporter, partial
(13%)
Length = 727
Score = 128 bits (321), Expect(2) = 2e-47
Identities = 59/133 (44%), Positives = 88/133 (65%)
Frame = +1
Query: 487 GMMSVAMTPNNHISIIVSSAFYSVWNLFSGFVLPRPGK**LISVILSIPVWWRWYSWANP 546
GM V++TPN ++ I+ S+FY++ NLFSG+ +PRP +P WW W + P
Sbjct: 226 GMFIVSVTPNVQLASILCSSFYTILNLFSGYFMPRP----------QMPKWWVWMYYLCP 375
Query: 547 VAWSLYGLVTSQYGDVKQNIETSDGRQTVEDFLRNYFGFKHDFLGVVALVNIAFPIVFAL 606
++W++ G++TSQYGD+ +N+ + +TV FL +Y+GF++DFLGV A+V I FPIVFA
Sbjct: 376 LSWAMNGMLTSQYGDINKNMSVFEDVKTVATFLEDYYGFRYDFLGVTAVVLIVFPIVFAA 555
Query: 607 VFAIAIKMFNFQR 619
+FA I NF R
Sbjct: 556 LFAYCIGNLNFPR 594
Score = 79.0 bits (193), Expect(2) = 2e-47
Identities = 35/71 (49%), Positives = 48/71 (67%)
Frame = +3
Query: 415 ERTVFYRERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGFEWTLVKVVWCLF 474
ERTV YRER A MYS + Y+ AQV+I++PY F+QA +Y I+ Y MIG W+ K+ W F
Sbjct: 9 ERTVLYRERFAGMYSPWAYSFAQVLIEVPYSFIQAALYVIITYPMIGHYWSAYKIFWSFF 188
Query: 475 FMYFTFLYFTF 485
M+ + L F +
Sbjct: 189 SMFCSILNFNY 221
>BP029778
Length = 438
Score = 177 bits (448), Expect = 6e-45
Identities = 87/124 (70%), Positives = 102/124 (82%)
Frame = -2
Query: 1 MVLPFEPHSITFDEVSYSVDMPQEMRNRGVIEDKLVLLKGLSGAFRPGVLTALMGVTGAG 60
M+LPF+ ++TF V+Y VDMPQE+R +G+IE KL LL +SG F PGVLTALMG +GAG
Sbjct: 386 MILPFQQFTVTFHNVNYYVDMPQEIRKQGIIETKLKLLSNVSGVFAPGVLTALMGSSGAG 207
Query: 61 KTTLMDVLSGRKTGGYIGGNITISGYPKKQETFARISGYCEQTDIHSPHVTVYESLLYSA 120
KTTLMDVL+GRKTGGYI G+I ISGYPK Q TFAR++GY EQ DIHSP VTV ESLL+SA
Sbjct: 206 KTTLMDVLAGRKTGGYIEGDIKISGYPKVQHTFARVAGYVEQNDIHSPQVTVEESLLFSA 27
Query: 121 WLRL 124
LRL
Sbjct: 26 LLRL 15
>AV408313
Length = 342
Score = 167 bits (422), Expect = 6e-42
Identities = 71/113 (62%), Positives = 92/113 (80%)
Frame = +3
Query: 280 VTTSSKEVELRIDYAEVYKNSELYRRNKALIKELSAPAPCSKDLYFPSRYSRSFFTQCIA 339
VT+S++EV + +D+ + YKNSELYRRNK LI EL PAP S DLYFP++YS+SF QC+A
Sbjct: 3 VTSSAQEVTIGVDFHQTYKNSELYRRNKQLIAELGIPAPGSNDLYFPTQYSQSFLVQCLA 182
Query: 340 CLWKQHWSYWRNPEYNAIRFLYSTAVAVLLGSMFWNLGSKIEKDQDLFNAMGS 392
CLWKQHWSYWRNP Y A+RF ++T +A++ G+MFW+LG K + QDLFNA+GS
Sbjct: 183 CLWKQHWSYWRNPPYTAVRFFFTTFIALIFGTMFWDLGGKYKNRQDLFNALGS 341
>CB828965
Length = 436
Score = 131 bits (329), Expect = 4e-31
Identities = 62/110 (56%), Positives = 75/110 (67%), Gaps = 1/110 (0%)
Frame = +1
Query: 511 WNLFSGFVLPRPGK**LISVILSIPVWWRWYSWANPVAWSLYGLVTSQYGDVKQNIETSD 570
WNLFSGF++PR IP+WWRWY WA+PVAW++YGLVTSQ GD IE
Sbjct: 7 WNLFSGFLIPRT----------QIPIWWRWYYWASPVAWTIYGLVTSQVGDKNSTIEVPG 156
Query: 571 GR-QTVEDFLRNYFGFKHDFLGVVALVNIAFPIVFALVFAIAIKMFNFQR 619
R TV+D+L FGF+HDFLGVVAL +IAF ++F VFA IK NFQ+
Sbjct: 157 FRPMTVKDYLERQFGFQHDFLGVVALTHIAFSLLFLFVFAYGIKFLNFQK 306
>TC9904 weakly similar to UP|Q8GU88 (Q8GU88) PDR-like ABC transporter,
partial (6%)
Length = 547
Score = 99.0 bits (245), Expect = 2e-21
Identities = 45/105 (42%), Positives = 63/105 (59%), Gaps = 2/105 (1%)
Frame = +1
Query: 516 GFVLPRPGK**LISVILSIPVWWRWYSWANPVAWSLYGLVTSQYGDVKQNIETS--DGRQ 573
GF +P P IP WW WY W PVAW++YGL+ S Y D+ + +
Sbjct: 1 GFFIPNP----------KIPKWWVWYYWICPVAWTVYGLIVSPYRDITPGLSVPGRSDQP 150
Query: 574 TVEDFLRNYFGFKHDFLGVVALVNIAFPIVFALVFAIAIKMFNFQ 618
++D++ +++GFK DF+G VA V +AFP+ FA VFA AIK+ NFQ
Sbjct: 151 ALKDYIEDHYGFKSDFMGPVACVLVAFPVFFAFVFAFAIKVLNFQ 285
>AW720269
Length = 524
Score = 87.4 bits (215), Expect = 6e-18
Identities = 55/158 (34%), Positives = 93/158 (58%), Gaps = 2/158 (1%)
Frame = +3
Query: 37 LLKGLSGAFRPGVLTALMGVTGAGKTTLMDVLSGR-KTGGYIGGNITISGYPKKQETFAR 95
+LK +S R + A++G +G GK+TL+ +++GR K + I+I+ +P R
Sbjct: 54 ILKSVSFVARSSEIVAVVGPSGTGKSTLLRIIAGRVKDRDFDPKTISINDHPMTSPAQLR 233
Query: 96 -ISGYCEQTDIHSPHVTVYESLLYSAWLRLSPDINAETRKMFIEEVMELVELKPLRYALV 154
I G+ Q D P +TV E+LL+SA RL ++ R+M +E +M+ + L + + V
Sbjct: 234 KICGFVAQEDNLLPLLTVKETLLFSAKFRLK-EMTPNDREMRVESLMQELGLFHVSDSFV 410
Query: 155 GLPGVSGLSTEQRKRLTVAVELVANPSIIFMDEPTSGL 192
G G+S +RKR+++ V+++ NP I+ +DEPTSGL
Sbjct: 411 GDEENRGISGGERKRVSIGVDMIHNPPILLLDEPTSGL 524
>AV416268
Length = 424
Score = 78.2 bits (191), Expect = 4e-15
Identities = 45/132 (34%), Positives = 80/132 (60%), Gaps = 3/132 (2%)
Frame = +3
Query: 110 VTVYESLLYSAWLRLSPDINAETRKMFIEEVMELVELKPLRYALVGLPGVSGLSTEQRKR 169
+TV E++ YSA L+L ++ ++ + ++ + L+ +G G G+S Q++R
Sbjct: 12 LTVGEAVYYSAHLQLPDSMSKSEKRERADFTIKEMGLQDAINTRIGGWGSKGVSGGQKRR 191
Query: 170 LTVAVELVANPSIIFMDEPTSGLDARAAAIVMRAVR--NTVD-TGRTVVCTIHQPSIDIF 226
+++ +E++ +P ++F+DEPTSGLD+ A+ V+ + N D RT+V +IHQPS +IF
Sbjct: 192 VSICIEILTHPRLLFLDEPTSGLDSAASYHVISRISSLNKKDGIQRTIVASIHQPSNEIF 371
Query: 227 ESFDELLLLKQG 238
+ F L LL G
Sbjct: 372 QLFHSLCLLSSG 407
>BP071638
Length = 406
Score = 74.3 bits (181), Expect = 5e-14
Identities = 34/41 (82%), Positives = 39/41 (94%)
Frame = -2
Query: 579 LRNYFGFKHDFLGVVALVNIAFPIVFALVFAIAIKMFNFQR 619
+R+YFGFKHDFLGVVA V +AFP+VFALVFAI+IKMFNFQR
Sbjct: 405 VRDYFGFKHDFLGVVAAVIVAFPVVFALVFAISIKMFNFQR 283
>BP049546
Length = 511
Score = 71.6 bits (174), Expect = 3e-13
Identities = 33/80 (41%), Positives = 49/80 (61%), Gaps = 1/80 (1%)
Frame = -1
Query: 541 YSWANPVAWSLYGLVTSQYGDVKQNIET-SDGRQTVEDFLRNYFGFKHDFLGVVALVNIA 599
Y WA+PVAWSL GL+TSQ G+ + DG TV+ FL+ G+ +DFL +A ++
Sbjct: 511 YYWASPVAWSLNGLITSQLGNKDAEVVIPGDGSMTVKQFLKENLGYHYDFLPAIAAAHVG 332
Query: 600 FPIVFALVFAIAIKMFNFQR 619
+ ++F FA IK N+Q+
Sbjct: 331 WVLLFGCGFAFGIKFLNYQK 272
>AV771159
Length = 486
Score = 70.1 bits (170), Expect = 1e-12
Identities = 40/106 (37%), Positives = 63/106 (58%)
Frame = -3
Query: 175 ELVANPSIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFESFDELLL 234
+L+ +PS++ +DEPTSGLD+ A ++ + GRTVV TIHQPS ++ FD+++L
Sbjct: 343 KLLIHPSLLLLDEPTSGLDSTTALRILNTIPRLATGGRTVVTTIHQPSSRLYYMFDKVVL 164
Query: 235 LKQGGQEIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEV 280
L + G IY GP +S + YF V + + NPA +L++
Sbjct: 163 LSE-GCPIYYGP----ASTALEYFSSVGFSTCVT--VNPADLLLDL 47
>BP055095
Length = 538
Score = 59.3 bits (142), Expect = 2e-09
Identities = 42/139 (30%), Positives = 73/139 (52%), Gaps = 1/139 (0%)
Frame = +3
Query: 161 GLSTEQRKRLTVAVELVANPSIIFMDEPTSGLDARAAAIVMRAVRNTVD-TGRTVVCTIH 219
G+S Q+KR+T LV +FMDE ++GLD+ + + +R V T+V ++
Sbjct: 99 GISGGQKKRVTTGEMLVGPAKALFMDEISTGLDSSTTFQICKFMRQMVHIMDVTMVISLL 278
Query: 220 QPSIDIFESFDELLLLKQGGQEIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLE 279
QP+ + FE FD+++LL + GQ +Y GP N++ +FE + K + A ++ E
Sbjct: 279 QPAPETFELFDDIILLSE-GQIVYQGP----RENVLEFFEYMG--FKCPERKGAADFLQE 437
Query: 280 VTTSSKEVELRIDYAEVYK 298
VT+ + + E Y+
Sbjct: 438 VTSKKDQEQYWFRKDEPYR 494
>TC18647 similar to UP|Q9ASR9 (Q9ASR9) At2g01320/F10A8.20, partial (17%)
Length = 430
Score = 57.8 bits (138), Expect = 5e-09
Identities = 35/104 (33%), Positives = 58/104 (55%), Gaps = 3/104 (2%)
Frame = +1
Query: 37 LLKGLSGAFRPGVLTALMGVTGAGKTTLMDVLSGRKTGG---YIGGNITISGYPKKQETF 93
LLK +SG +PG L A+MG +G+GKTTL++VL+G+ ++ G + +G P + +
Sbjct: 106 LLKNVSGEAKPGRLLAIMGPSGSGKTTLLNVLAGQLAASPRLHLSGLLEFNGKPGSKNAY 285
Query: 94 ARISGYCEQTDIHSPHVTVYESLLYSAWLRLSPDINAETRKMFI 137
Y Q D+ +TV E+L + L+L +AE R ++
Sbjct: 286 K--FAYVRQEDLFFSQLTVRETLSLATELQLPNISSAEERDEYV 411
>TC11425 similar to UP|Q9M1Q9 (Q9M1Q9) P-glycoprotein-like proetin, partial
(9%)
Length = 712
Score = 45.8 bits (107), Expect = 2e-05
Identities = 37/123 (30%), Positives = 61/123 (49%)
Frame = +2
Query: 162 LSTEQRKRLTVAVELVANPSIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQP 221
LS Q++R+ +A ++ P+I+ +DE TS LDA + +V A+ + V RT V H+
Sbjct: 107 LSGGQKQRVAIARAIIKTPNILLLDEATSALDAESERVVQDAL-DKVMVNRTTVIVAHR- 280
Query: 222 SIDIFESFDELLLLKQGGQEIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVT 281
+ ++ D + +LK G + V H + LIN IKDGY + L T
Sbjct: 281 -LSTIKNADVITVLKNG---VIVEKGRHET--LIN----------IKDGYYASLVQLHTT 412
Query: 282 TSS 284
++
Sbjct: 413 ATT 421
>BE122583
Length = 361
Score = 40.0 bits (92), Expect(2) = 6e-05
Identities = 15/29 (51%), Positives = 23/29 (78%)
Frame = +3
Query: 1 MVLPFEPHSITFDEVSYSVDMPQEMRNRG 29
MVLPF+P S+ F+ V+Y ++MP EM+ +G
Sbjct: 180 MVLPFQPLSLAFENVNYYIEMPNEMKKQG 266
Score = 23.5 bits (49), Expect(2) = 6e-05
Identities = 10/17 (58%), Positives = 13/17 (75%)
Frame = +1
Query: 45 FRPGVLTALMGVTGAGK 61
F+ TAL+GV+GAGK
Sbjct: 310 FQASDFTALVGVSGAGK 360
>TC10750 similar to UP|AAP80385 (AAP80385) ABC transporter, partial (38%)
Length = 807
Score = 44.3 bits (103), Expect = 6e-05
Identities = 47/223 (21%), Positives = 85/223 (38%), Gaps = 1/223 (0%)
Frame = +1
Query: 332 SFFTQCIACLWKQHWSYWRNPEYNAIRFLYSTAVAVLLGSMFWNLGSKIEKDQDLFNAMG 391
SF Q + + R+ Y +R + V + +G+++ N+G+ A G
Sbjct: 139 SFLMQSYTLTKRSFINMSRDFGYYWLRLVIYIVVTICIGTIYLNVGTGYNS----ILARG 306
Query: 392 SMYAAVILIGAMNSNSVQPVVGVERTVFYRERAARMYSAFPYALAQVVIQLPYVFVQAVV 451
S + V S P + VF RER Y + ++ V P++ + +
Sbjct: 307 SCASFVFGFVTFMSIGGFPSFVEDMKVFQRERLNGHYGVTAFVVSNTVSATPFLVLITFL 486
Query: 452 YGIVVYVMIGFEWTLVKVVWCLFFMYFTFLYFTFYGMMSVAMTPNNHISIIVSSAFYSVW 511
G + Y M+ V+ + +Y + M ++ PN + II+ + ++
Sbjct: 487 SGTICYFMVRLHPGFSHYVFFVLCLYASVTVVESLMMAIASIVPNFLMGIIIGAGIQGIF 666
Query: 512 NLFSG-FVLPRPGK**LISVILSIPVWWRWYSWANPVAWSLYG 553
L SG F LP + PVW S+ + W+L G
Sbjct: 667 MLVSGYFRLPHD---------IPKPVWRYPMSYISFHFWALQG 768
>TC15722 similar to UP|Q9FMU0 (Q9FMU0) Similarity to ABC transporter,
partial (37%)
Length = 624
Score = 42.7 bits (99), Expect = 2e-04
Identities = 22/59 (37%), Positives = 38/59 (64%), Gaps = 6/59 (10%)
Frame = +3
Query: 154 VGLPGVS------GLSTEQRKRLTVAVELVANPSIIFMDEPTSGLDARAAAIVMRAVRN 206
VGL G+S LS ++RL +A++LV P ++ +DEP +GLD +A A V++ +++
Sbjct: 27 VGLSGISLDKNPQSLSGGYKRRLALAIQLVQVPDLLILDEPLAGLDWKARADVVKLLKH 203
>BP042918
Length = 412
Score = 42.0 bits (97), Expect = 3e-04
Identities = 28/145 (19%), Positives = 61/145 (41%), Gaps = 5/145 (3%)
Frame = -3
Query: 418 VFYRERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGFEWTLVKVVWCLFFMY 477
VFY+ R + A+ Y + ++++P +++V+ + Y GF + L ++
Sbjct: 392 VFYKHRDHLFHPAWTYTVPNFLLRIPISIFESLVWVAITYYTTGFAPEASRFFKQLLVVF 213
Query: 478 F-----TFLYFTFYGMMSVAMTPNNHISIIVSSAFYSVWNLFSGFVLPRPGK**LISVIL 532
++ G+ + N ++++ F L GF+LP+
Sbjct: 212 LIQQMAAGMFRLISGVCRTMIIANTGGALMLLLVF-----LLGGFLLPKR---------- 78
Query: 533 SIPVWWRWYSWANPVAWSLYGLVTS 557
+IP WW W W +P++++ L +
Sbjct: 77 AIPDWWVWAYWISPLSYAFNSLTVN 3
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.326 0.140 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,588,207
Number of Sequences: 28460
Number of extensions: 157614
Number of successful extensions: 1495
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 1454
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1479
length of query: 619
length of database: 4,897,600
effective HSP length: 96
effective length of query: 523
effective length of database: 2,165,440
effective search space: 1132525120
effective search space used: 1132525120
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148607.16


