

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148532.8 + phase: 0
(380 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
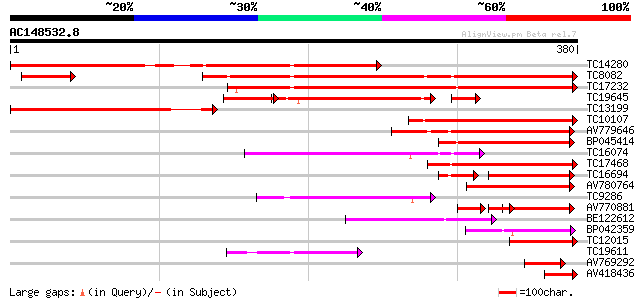
Score E
Sequences producing significant alignments: (bits) Value
TC14280 weakly similar to UP|Q94IC5 (Q94IC5) BURP domain-contain... 286 4e-78
TC8082 similar to UP|Q8H9E8 (Q8H9E8) Resistant specific protein-... 235 8e-63
TC17232 weakly similar to UP|Q8H9E9 (Q8H9E9) Resistant specific ... 232 7e-62
TC19645 similar to UP|Q94IC5 (Q94IC5) BURP domain-containing pro... 152 7e-55
TC13199 similar to UP|Q94IC5 (Q94IC5) BURP domain-containing pro... 191 2e-49
TC10107 similar to UP|Q9MB24 (Q9MB24) S1-3 protein (Fragment), p... 186 6e-48
AV779646 153 5e-38
BP045414 120 4e-28
TC16074 similar to UP|Q8H9E8 (Q8H9E8) Resistant specific protein... 115 2e-26
TC17468 weakly similar to UP|Q8H9E9 (Q8H9E9) Resistant specific ... 105 1e-23
TC16694 similar to UP|Q8H9E9 (Q8H9E9) Resistant specific protein... 90 7e-23
AV780764 91 4e-19
TC9286 similar to UP|O65009 (O65009) BURP domain containing prot... 86 1e-17
AV770881 61 4e-16
BE122612 74 5e-14
BP042359 72 1e-13
TC12015 weakly similar to UP|O65009 (O65009) BURP domain contain... 64 5e-11
TC19611 61 3e-10
AV769292 52 1e-07
AV418436 47 5e-06
>TC14280 weakly similar to UP|Q94IC5 (Q94IC5) BURP domain-containing protein
(Fragment), partial (54%)
Length = 723
Score = 286 bits (732), Expect = 4e-78
Identities = 153/250 (61%), Positives = 186/250 (74%), Gaps = 1/250 (0%)
Frame = +2
Query: 1 MCVYVVLQLALVAT-HAALPPEVYWKSKLPTTQMPKAITDLLHLDWTEEKSTSVAVGKGG 59
+ V +L + LVAT HAALPPE+YWKS LPTT MPKAITD+L+ W E+KST+V V KGG
Sbjct: 29 LSVCALLSVLLVATSHAALPPELYWKSVLPTTPMPKAITDILYPHWVEDKSTNVGVRKGG 208
Query: 60 VNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRVNV 119
VNVD+GK KPG T V+VGKGGV+V+AGK G G GG +VN G GK
Sbjct: 209 VNVDAGKGKPGHTKVDVGKGGVNVNAGKGSGG------GHGGTNVNVGGGK--------- 343
Query: 120 GKGGVNVHAGKGRGKPVHVSVGNKSPFLYNYAASETQLHDKPNVALFFLEKDLHQGTKLN 179
GGV+VH G +GKPV V+VG+KSPF Y YAA+ETQLH PNVALFFLEK+LH GTK+N
Sbjct: 344 -GGGVSVHTGH-KGKPVFVNVGSKSPFDYVYAATETQLHSDPNVALFFLEKNLHPGTKMN 517
Query: 180 LQFTKTNSNDDAKFLPKEVANSIPFSSSKVENILNLFSIKEGTEESEIVKNTISECEENG 239
L FTKT++ A FLP+ VA+SIPFSS KVE LN +S+K G+EE++++KNTI ECE+ G
Sbjct: 518 LHFTKTSNK--ATFLPRIVADSIPFSSEKVELALNKYSVKAGSEEAQVMKNTIDECEQKG 691
Query: 240 IKGEEKLCVT 249
I+GEEK C T
Sbjct: 692 IQGEEKYCAT 721
>TC8082 similar to UP|Q8H9E8 (Q8H9E8) Resistant specific protein-3, partial
(54%)
Length = 1913
Score = 235 bits (600), Expect = 8e-63
Identities = 118/251 (47%), Positives = 163/251 (64%)
Frame = +1
Query: 130 KGRGKPVHVSVGNKSPFLYNYAASETQLHDKPNVALFFLEKDLHQGTKLNLQFTKTNSND 189
+ + P+ + VG +S N AS+ LH+ FF E LH K + K S+
Sbjct: 892 QSQDNPLRIYVGKESQH-ENSGASDHHLHEISARVGFFQEDQLHSDNKFYMHLNKIRSS- 1065
Query: 190 DAKFLPKEVANSIPFSSSKVENILNLFSIKEGTEESEIVKNTISECEENGIKGEEKLCVT 249
LP+++ IPFS K++ +L + S+K ++ +EI++ TIS CE + GEEK C T
Sbjct: 1066 -IPLLPRQITQQIPFSLDKMKEVLEMLSVKPNSKNAEIMEKTISGCEAPAMDGEEKYCAT 1242
Query: 250 SLESMVDFTTSKLGNNVEAVSTEVKKESSDLQEYVMAKGVKKLGEKNKAVVCHKESYPYA 309
SLESM+DF TSKLG NV A+ST+V+KE+ LQ ++ GVKKL + + + CH +YPY
Sbjct: 1243 SLESMMDFVTSKLGKNVHAMSTDVQKETK-LQNLLVKDGVKKLADDH-VIACHPLAYPYV 1416
Query: 310 VFYCHKTDSTKVYSVPLEGVDGSRVKAVAVCHTDTSQWNPKHLAFQVLNVQPGTVPVCHF 369
VF CH+ T Y VPLEG DG RVKAVAVCH DTS+W+P ++AF+VL V+PGTVPVCHF
Sbjct: 1417 VFGCHELQKTSAYFVPLEGEDGVRVKAVAVCHKDTSKWDPNYVAFRVLKVKPGTVPVCHF 1596
Query: 370 LPQDHVVWVSK 380
P+ H++W+SK
Sbjct: 1597 FPEGHLLWLSK 1629
Score = 52.8 bits (125), Expect = 1e-07
Identities = 22/36 (61%), Positives = 28/36 (77%)
Frame = +1
Query: 9 LALVATHAALPPEVYWKSKLPTTQMPKAITDLLHLD 44
+ L+ THAALPPE+YWK LP+T MPKA+ +LL D
Sbjct: 70 MMLMTTHAALPPEIYWKMMLPSTPMPKAVIELLRGD 177
>TC17232 weakly similar to UP|Q8H9E9 (Q8H9E9) Resistant specific protein-2,
partial (33%)
Length = 966
Score = 232 bits (592), Expect = 7e-62
Identities = 118/238 (49%), Positives = 160/238 (66%), Gaps = 4/238 (1%)
Frame = +3
Query: 147 LYNY---AASETQLHDKPNVALFFLEKDLHQGTKLNLQFTKTNSNDDAKFLPKEVANSIP 203
LY Y A+ LH+K +FF E++LH GTKL+LQ +K LP+++A+ IP
Sbjct: 69 LYGYKSGASLHKLLHEKTKAGVFFFEENLHPGTKLDLQLSKRKPA--VPILPRKIADQIP 242
Query: 204 FSSSKVENILNLFSIKEGTEESEIVKNTISECEENGIKGEEKLCVTSLESMVDFTTSKLG 263
FS K++ IL + +K ++ +EIV+ T+ CE ++GEE++C TSLES+VDF SKLG
Sbjct: 243 FSLEKLKEILEILVVKPDSKNTEIVEKTMGGCELPSMEGEEQMCATSLESLVDFIASKLG 422
Query: 264 NNVEAVSTEVKKESSDLQEYVMAK-GVKKLGEKNKAVVCHKESYPYAVFYCHKTDSTKVY 322
NV A+STEV+ E + ++ K GVKKL + AV CH +YPY V + HK T Y
Sbjct: 423 KNVLAISTEVELEKETQSQNLLVKDGVKKLADDGMAV-CHPMTYPYPVCFSHKLVKTSAY 599
Query: 323 SVPLEGVDGSRVKAVAVCHTDTSQWNPKHLAFQVLNVQPGTVPVCHFLPQDHVVWVSK 380
VPLEG DG+RVKAVAVCH DTS+W+P H++F LNV+PGTVPVCH LP+ H++WV K
Sbjct: 600 FVPLEGEDGARVKAVAVCHRDTSKWDPNHVSFPDLNVKPGTVPVCHVLPERHLLWVPK 773
>TC19645 similar to UP|Q94IC5 (Q94IC5) BURP domain-containing protein
(Fragment), partial (38%)
Length = 619
Score = 152 bits (383), Expect(3) = 7e-55
Identities = 81/112 (72%), Positives = 97/112 (86%), Gaps = 2/112 (1%)
Frame = +3
Query: 176 TKLNLQFTKTNSNDDAK--FLPKEVANSIPFSSSKVENILNLFSIKEGTEESEIVKNTIS 233
TKLNLQFTKT SND A FLP++VA+SIPFSS+KV+++LN FS+K G+EE+EIVKNTI+
Sbjct: 120 TKLNLQFTKT-SNDAAATFFLPRQVADSIPFSSNKVDDVLNKFSLKPGSEEAEIVKNTIA 296
Query: 234 ECEENGIKGEEKLCVTSLESMVDFTTSKLGNNVEAVSTEVKKESSDLQEYVM 285
ECEE G+ GEEK CVTSLESMVDFTTSKLG NVEAVST+V KE +LQ+Y +
Sbjct: 297 ECEEPGVVGEEKRCVTSLESMVDFTTSKLGKNVEAVSTDVNKE-KELQQYTI 449
Score = 68.2 bits (165), Expect(3) = 7e-55
Identities = 31/37 (83%), Positives = 33/37 (88%)
Frame = +2
Query: 144 SPFLYNYAASETQLHDKPNVALFFLEKDLHQGTKLNL 180
SPF YNYAASETQLH+ PNVALFFLEKDLHQG K+ L
Sbjct: 23 SPFDYNYAASETQLHENPNVALFFLEKDLHQGNKIEL 133
Score = 31.6 bits (70), Expect(3) = 7e-55
Identities = 12/19 (63%), Positives = 15/19 (78%)
Frame = +2
Query: 297 KAVVCHKESYPYAVFYCHK 315
K ++ HKE+YPYAVF C K
Sbjct: 482 KLLLFHKENYPYAVFXCXK 538
>TC13199 similar to UP|Q94IC5 (Q94IC5) BURP domain-containing protein
(Fragment), partial (19%)
Length = 426
Score = 191 bits (484), Expect = 2e-49
Identities = 95/139 (68%), Positives = 111/139 (79%)
Frame = +2
Query: 1 MCVYVVLQLALVATHAALPPEVYWKSKLPTTQMPKAITDLLHLDWTEEKSTSVAVGKGGV 60
+ ++++L LALVAT A+L PE+YWKSKLPTT MPKAITDLLH DWTE+KSTSVAVGKGGV
Sbjct: 68 LSIFMLLNLALVATQASLTPELYWKSKLPTTPMPKAITDLLHSDWTEDKSTSVAVGKGGV 247
Query: 61 NVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRVNVG 120
NVD+GKTKPGGT+VNVGKGGVHV+AGK KPGGT+VNVGKGGV+V+AGK
Sbjct: 248 NVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGTSVNVGKGGVNVHAGK------------ 391
Query: 121 KGGVNVHAGKGRGKPVHVS 139
GKG+ KPVHV+
Sbjct: 392 --------GKGKRKPVHVN 424
>TC10107 similar to UP|Q9MB24 (Q9MB24) S1-3 protein (Fragment), partial
(65%)
Length = 575
Score = 186 bits (472), Expect = 6e-48
Identities = 88/113 (77%), Positives = 99/113 (86%)
Frame = +2
Query: 268 AVSTEVKKESSDLQEYVMAKGVKKLGEKNKAVVCHKESYPYAVFYCHKTDSTKVYSVPLE 327
AVST V KE+ + Q Y +A GVKKL +KAVVCHK++YPYAVFYCHK+DST+ YSVPLE
Sbjct: 2 AVSTVVDKET-EKQRYTIATGVKKLEAGDKAVVCHKQNYPYAVFYCHKSDSTRAYSVPLE 178
Query: 328 GVDGSRVKAVAVCHTDTSQWNPKHLAFQVLNVQPGTVPVCHFLPQDHVVWVSK 380
G +G RVKAVAVCHTDTSQWNPKHLAFQVL V+PGTVPVCHFLP+DHVVWVSK
Sbjct: 179 GDNGVRVKAVAVCHTDTSQWNPKHLAFQVLKVKPGTVPVCHFLPEDHVVWVSK 337
>AV779646
Length = 510
Score = 153 bits (386), Expect = 5e-38
Identities = 75/123 (60%), Positives = 92/123 (73%), Gaps = 1/123 (0%)
Frame = -3
Query: 257 FTTSKLGNNVEAVSTEVKKESSDLQEYVMAK-GVKKLGEKNKAVVCHKESYPYAVFYCHK 315
F TSKLGN+V A+STEV KE+ L Y++ K GV+KL + + VCH +YPY VF+CHK
Sbjct: 508 FITSKLGNDVHAMSTEVTKETKLL--YLLVKDGVEKLAD-DYITVCHPMAYPYVVFWCHK 338
Query: 316 TDSTKVYSVPLEGVDGSRVKAVAVCHTDTSQWNPKHLAFQVLNVQPGTVPVCHFLPQDHV 375
T Y VPLEG DG RVKAVAVCH DTS+W+P H AFQ L V+PGTVPVCHFLP+ H+
Sbjct: 337 LQKTDAYFVPLEGEDGVRVKAVAVCHKDTSKWDPNHAAFQDLKVKPGTVPVCHFLPEGHL 158
Query: 376 VWV 378
+W+
Sbjct: 157 LWL 149
>BP045414
Length = 478
Score = 120 bits (301), Expect = 4e-28
Identities = 56/91 (61%), Positives = 68/91 (74%)
Frame = -2
Query: 288 GVKKLGEKNKAVVCHKESYPYAVFYCHKTDSTKVYSVPLEGVDGSRVKAVAVCHTDTSQW 347
GVKKL + AV CH +YPYAVF+CHK T Y VPLEG DG+RV VAVC DTS+W
Sbjct: 471 GVKKLPDDGIAV-CHPLTYPYAVFFCHKLVKTSAYFVPLEGEDGARV*PVAVCPRDTSKW 295
Query: 348 NPKHLAFQVLNVQPGTVPVCHFLPQDHVVWV 378
+P H++F L+VQPGTVPVCH LP+ H++WV
Sbjct: 294 DPNHVSFPDLHVQPGTVPVCHVLPERHLLWV 202
>TC16074 similar to UP|Q8H9E8 (Q8H9E8) Resistant specific protein-3, partial
(6%)
Length = 844
Score = 115 bits (287), Expect = 2e-26
Identities = 62/164 (37%), Positives = 96/164 (57%), Gaps = 3/164 (1%)
Frame = +2
Query: 158 HDKPNVALFFLEKDLHQGTKLNLQFTKTNSNDDAKFLPKEVANSIPFSSSKVENILNLFS 217
H P + +FF+ KDL G K+ + F+ +S+ KFLP+E AN IPFSS ++ ++LN FS
Sbjct: 353 HMDPELHVFFIPKDLKVGKKMPIYFSMKDSSTSPKFLPREEANQIPFSSKQLPSLLNFFS 532
Query: 218 IKEGTEESEIVKNTISECEENGIKGEEKLCVTSLESMVDFTTSKLGNNVE---AVSTEVK 274
I + + +++ +K T+ +CE ++GE K C TSLES++D G + + +T +
Sbjct: 533 IPKNSPQAKAMKYTLEQCEFEPMEGETKFCATSLESLLDSARHLFGLSPQFKVLTTTHLT 712
Query: 275 KESSDLQEYVMAKGVKKLGEKNKAVVCHKESYPYAVFYCHKTDS 318
K LQ Y + + VK+ N + CH YP+AVFYCH S
Sbjct: 713 KSHVLLQNYTILE-VKENRVPN-VIGCHPMPYPFAVFYCHSQQS 838
>TC17468 weakly similar to UP|Q8H9E9 (Q8H9E9) Resistant specific protein-2,
partial (11%)
Length = 652
Score = 105 bits (262), Expect = 1e-23
Identities = 46/101 (45%), Positives = 64/101 (62%), Gaps = 1/101 (0%)
Frame = +3
Query: 281 QEYVMAKGVKKLGEKNKAVVCHKESYPYAVFYCHKTDS-TKVYSVPLEGVDGSRVKAVAV 339
Q Y + + + ++ K V CH YPYAVFYCH +S +VY V L G +G +V+A+ V
Sbjct: 3 QNYTILENIMRISAP-KMVACHTMPYPYAVFYCHSQESENRVYKVSLVGDNGDKVEAMVV 179
Query: 340 CHTDTSQWNPKHLAFQVLNVQPGTVPVCHFLPQDHVVWVSK 380
CH DTS W H++FQ+L V PG+ VCHF P D+++W K
Sbjct: 180 CHLDTSHWGSGHVSFQILGVTPGSSSVCHFFPADNLIWAPK 302
>TC16694 similar to UP|Q8H9E9 (Q8H9E9) Resistant specific protein-2, partial
(12%)
Length = 488
Score = 89.7 bits (221), Expect(2) = 7e-23
Identities = 38/57 (66%), Positives = 45/57 (78%)
Frame = +1
Query: 322 YSVPLEGVDGSRVKAVAVCHTDTSQWNPKHLAFQVLNVQPGTVPVCHFLPQDHVVWV 378
Y VPLEG DG RV VAVCH DTS+W+P H AF L V+PGTVPVCHFLP+ H++W+
Sbjct: 106 YFVPLEGEDGVRVNHVAVCHQDTSKWDPNHAAFPDLKVKPGTVPVCHFLPEGHLLWL 276
Score = 33.9 bits (76), Expect(2) = 7e-23
Identities = 14/27 (51%), Positives = 19/27 (69%)
Frame = +3
Query: 288 GVKKLGEKNKAVVCHKESYPYAVFYCH 314
GV+KL + + VCH +YPY VF+CH
Sbjct: 6 GVEKLAD-DYITVCHPMAYPYLVFWCH 83
>AV780764
Length = 438
Score = 90.5 bits (223), Expect = 4e-19
Identities = 39/72 (54%), Positives = 49/72 (67%)
Frame = -1
Query: 307 PYAVFYCHKTDSTKVYSVPLEGVDGSRVKAVAVCHTDTSQWNPKHLAFQVLNVQPGTVPV 366
PY VF+ H + Y VPLEG DG RVK +AVCH TS+W+P H A L V+PGT PV
Sbjct: 378 PYVVFWFH*LPKPQAYFVPLEGEDGVRVKTIAVCHQHTSKWDPNHAALPNLKVKPGTGPV 199
Query: 367 CHFLPQDHVVWV 378
CHFL + H++W+
Sbjct: 198 CHFLLEGHLLWL 163
>TC9286 similar to UP|O65009 (O65009) BURP domain containing protein,
partial (6%)
Length = 805
Score = 85.5 bits (210), Expect = 1e-17
Identities = 46/123 (37%), Positives = 73/123 (58%), Gaps = 3/123 (2%)
Frame = +1
Query: 166 FFLEKDLHQGTKLNLQFTKTNSNDDAKFLPKEVANSIPFSSSKVENILNLFSIKEGTEES 225
FF DL+ G + LQF + + FL ++ N IPFSSS++ ++L LFSI E + ++
Sbjct: 445 FFALDDLYAGNVMTLQFPV---QEISHFLSRKETNYIPFSSSQLPSVLQLFSIPEDSPKA 615
Query: 226 EIVKNTISECEENGIKGEEKLCVTSLESMVDFTTSKLGNNVEA---VSTEVKKESSDLQE 282
+ ++ T+ +CE I GE K+C TSLESM++F S +G+N E ++ S LQ+
Sbjct: 616 KSMRGTLGQCEGEPITGERKICATSLESMLEFVDSVIGSNTEGDLLTTSHPSPSGSPLQK 795
Query: 283 YVM 285
Y +
Sbjct: 796 YTI 804
>AV770881
Length = 403
Score = 60.8 bits (146), Expect(3) = 4e-16
Identities = 25/48 (52%), Positives = 34/48 (70%)
Frame = -3
Query: 331 GSRVKAVAVCHTDTSQWNPKHLAFQVLNVQPGTVPVCHFLPQDHVVWV 378
G +K + VCH DTS+W+P H AF L V+PGTVPV FLP+ ++W+
Sbjct: 311 GLGLKPLLVCHQDTSKWDPNHAAFPDLKVKPGTVPVSQFLPEGPLLWL 168
Score = 30.0 bits (66), Expect(3) = 4e-16
Identities = 10/19 (52%), Positives = 12/19 (62%)
Frame = -1
Query: 301 CHKESYPYAVFYCHKTDST 319
CH +YPY VF+CH T
Sbjct: 403 CHPMAYPYGVFWCHGLPKT 347
Score = 29.6 bits (65), Expect(3) = 4e-16
Identities = 14/17 (82%), Positives = 14/17 (82%)
Frame = -2
Query: 322 YSVPLEGVDGSRVKAVA 338
Y VPLEG DG RVKAVA
Sbjct: 339 YFVPLEGEDGVRVKAVA 289
>BE122612
Length = 386
Score = 73.6 bits (179), Expect = 5e-14
Identities = 37/101 (36%), Positives = 57/101 (55%)
Frame = +2
Query: 226 EIVKNTISECEENGIKGEEKLCVTSLESMVDFTTSKLGNNVEAVSTEVKKESSDLQEYVM 285
+++++++ ECE +GE K CV S+E MVDF TS LG NV +TE S+
Sbjct: 35 KMMRDSLQECERVPSRGETKRCVGSIEDMVDFATSVLGRNVVVRTTENVNGSNKKVMVGR 214
Query: 286 AKGVKKLGEKNKAVVCHKESYPYAVFYCHKTDSTKVYSVPL 326
+GV G+ ++V CH+ +PY ++YCH +VY L
Sbjct: 215 VQGVNG-GKVTRSVSCHQSLFPYLLYYCHSLPKVRVYQADL 334
>BP042359
Length = 404
Score = 72.4 bits (176), Expect = 1e-13
Identities = 31/77 (40%), Positives = 46/77 (59%), Gaps = 3/77 (3%)
Frame = -3
Query: 306 YPYAVFYCHKTDSTKVYSVPLEGVDGSRVK---AVAVCHTDTSQWNPKHLAFQVLNVQPG 362
YPY ++YCH +VY + VD +++K VA+CH DTS W P+H AF L PG
Sbjct: 402 YPYLLYYCHSVPQVRVYEAEILDVD-TKLKINHGVAICHLDTSAWGPEHGAFVALGAGPG 226
Query: 363 TVPVCHFLPQDHVVWVS 379
+ VCH++ Q+ + W +
Sbjct: 225 KIEVCHWIFQNDMTWTT 175
>TC12015 weakly similar to UP|O65009 (O65009) BURP domain containing
protein, partial (12%)
Length = 464
Score = 63.5 bits (153), Expect = 5e-11
Identities = 23/45 (51%), Positives = 31/45 (68%)
Frame = +1
Query: 336 AVAVCHTDTSQWNPKHLAFQVLNVQPGTVPVCHFLPQDHVVWVSK 380
A+ VCH DTS W P H+ F+ L ++ G PVCHF P H++WV+K
Sbjct: 1 ALGVCHLDTSDWTPSHIIFKQLGIEAGKAPVCHFFPIKHLMWVAK 135
>TC19611
Length = 533
Score = 61.2 bits (147), Expect = 3e-10
Identities = 35/91 (38%), Positives = 49/91 (53%)
Frame = -3
Query: 146 FLYNYAASETQLHDKPNVALFFLEKDLHQGTKLNLQFTKTNSNDDAKFLPKEVANSIPFS 205
F Y A ETQ FF E LH G K N+ K S +LP+E+A+ IPFS
Sbjct: 258 FYAYYVAKETQSRS------FFEEYQLHAGNKFNVNLNKIKSV--VPWLPREIADHIPFS 103
Query: 206 SSKVENILNLFSIKEGTEESEIVKNTISECE 236
K+ IL + S+K ++ +E+V+ TI +CE
Sbjct: 102 LDKMNEILEMLSVKLESKNAEMVEKTIGDCE 10
Score = 38.1 bits (87), Expect = 0.002
Identities = 19/40 (47%), Positives = 24/40 (59%)
Frame = -2
Query: 11 LVATHAALPPEVYWKSKLPTTQMPKAITDLLHLDWTEEKS 50
++ ALP E+ WK KLP T +PKAIT+L D KS
Sbjct: 466 VMTAQGALPAELDWKRKLPATPVPKAITEL*RGDVNTGKS 347
>AV769292
Length = 250
Score = 52.4 bits (124), Expect = 1e-07
Identities = 22/27 (81%), Positives = 24/27 (88%)
Frame = -3
Query: 346 QWNPKHLAFQVLNVQPGTVPVCHFLPQ 372
+WN KHLAFQVL V+PGTV VCHFLPQ
Sbjct: 248 KWNLKHLAFQVLKVKPGTVSVCHFLPQ 168
>AV418436
Length = 280
Score = 47.0 bits (110), Expect = 5e-06
Identities = 19/22 (86%), Positives = 20/22 (90%)
Frame = -1
Query: 359 VQPGTVPVCHFLPQDHVVWVSK 380
V+PGTV VCHFLPQDHVVWV K
Sbjct: 277 VKPGTVSVCHFLPQDHVVWVPK 212
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.312 0.131 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,371,173
Number of Sequences: 28460
Number of extensions: 85652
Number of successful extensions: 471
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 419
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 440
length of query: 380
length of database: 4,897,600
effective HSP length: 92
effective length of query: 288
effective length of database: 2,279,280
effective search space: 656432640
effective search space used: 656432640
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC148532.8


