

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148528.7 - phase: 0 /pseudo
(1391 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
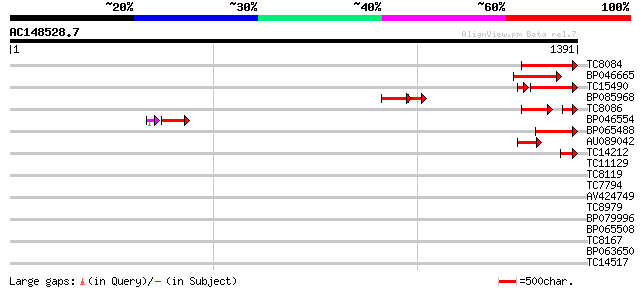
Score E
Sequences producing significant alignments: (bits) Value
TC8084 similar to UP|PIF1_YEAST (P07271) DNA repair and recombin... 158 6e-39
BP046665 150 2e-36
TC15490 weakly similar to UP|Q9LTU4 (Q9LTU4) Helicase-like prote... 120 5e-33
BP085968 85 9e-25
TC8086 similar to UP|DMC1_HUMAN (Q14565) Meiotic recombination p... 85 3e-24
BP046554 74 2e-17
BP065488 80 3e-15
AU089042 62 5e-10
TC14212 similar to UP|Q9FN61 (Q9FN61) Gb|AAF07369.1, partial (10%) 42 9e-04
TC11129 weakly similar to UP|PDA6_MEDSA (P38661) Probable protei... 37 0.016
TC8119 similar to UP|Q9FFF5 (Q9FFF5) Nucleic acid binding protei... 31 1.2
TC7794 homologue to UP|Q9M600 (Q9M600) Chlorophyll a/b binding p... 30 2.0
AV424749 30 2.0
TC8979 weakly similar to UP|Q8RY59 (Q8RY59) At1g32230/F3C3_1, pa... 30 2.6
BP079996 29 5.8
BP065508 29 5.8
TC8167 similar to PIR|JC7519|JC7519 subtilisin-like serine prote... 28 7.6
BP063650 28 7.6
TC14517 28 9.9
>TC8084 similar to UP|PIF1_YEAST (P07271) DNA repair and recombination
protein PIF1, mitochondrial precursor, partial (4%)
Length = 560
Score = 158 bits (399), Expect = 6e-39
Identities = 77/135 (57%), Positives = 102/135 (75%)
Frame = +1
Query: 1257 LDQVEGLCNGTRLIVTRMANHVIEAKIISGKNVGNLTYIPRMSMSPSQSPWPFKLIRRQF 1316
+ + E LCNGTRLIV + +VI+A +I+G N+G+ +IPR+ M PS S +PFK RR F
Sbjct: 43 IQKFENLCNGTRLIVVDLGTYVIKATVITGTNIGDDIFIPRLDMVPSDSGYPFKFERR*F 222
Query: 1317 PIIVSYAMTINKSQGQTLDSVGLYLPRSVFSHGQLYVAFSRVRTKAGLKILIHDLEKKPL 1376
PI + +AMTINKSQGQ+L V LYL R VF+HGQLYVA SRVR++ GLK+L+ D E+K
Sbjct: 223 PISLCFAMTINKSQGQSLSHVSLYLSRPVFTHGQLYVALSRVRSRKGLKLLVLDEEEKVT 402
Query: 1377 SNTTNVVYKEVFDNL 1391
+ T NVVY+EVF+N+
Sbjct: 403 NTTKNVVYREVFENI 447
>BP046665
Length = 524
Score = 150 bits (378), Expect = 2e-36
Identities = 75/118 (63%), Positives = 89/118 (74%)
Frame = -1
Query: 1237 GLPNHKIKLKVGTPIMLLRNLDQVEGLCNGTRLIVTRMANHVIEAKIISGKNVGNLTYIP 1296
G+PNHKI LK G PIMLLRN+ Q G CNGTRLIV + +VI+A +I+ N+G+ +IP
Sbjct: 509 GIPNHKITLKEGAPIMLLRNIYQAVGFCNGTRLIVADLGTNVIKATVIT*TNIGDDIFIP 330
Query: 1297 RMSMSPSQSPWPFKLIRRQFPIIVSYAMTINKSQGQTLDSVGLYLPRSVFSHGQLYVA 1354
RM M PS S +PFK RRQFPI + AMTINKSQGQ+L VGLYL R VF+HGQLYVA
Sbjct: 329 RMDMVPSDSGYPFKFERRQFPISLCSAMTINKSQGQSLSHVGLYLSRHVFTHGQLYVA 156
Score = 35.0 bits (79), Expect = 0.081
Identities = 17/45 (37%), Positives = 28/45 (61%)
Frame = -2
Query: 1340 YLPRSVFSHGQLYVAFSRVRTKAGLKILIHDLEKKPLSNTTNVVY 1384
Y+ +++SH S +R++ GLK+L+ D E+K + T NVVY
Sbjct: 202 YIFLAMYSHMVSCTLLSMLRSRKGLKLLVLDEEEKVTNTTKNVVY 68
>TC15490 weakly similar to UP|Q9LTU4 (Q9LTU4) Helicase-like protein, partial
(8%)
Length = 634
Score = 120 bits (300), Expect(2) = 5e-33
Identities = 60/113 (53%), Positives = 82/113 (72%)
Frame = +2
Query: 1279 IEAKIISGKNVGNLTYIPRMSMSPSQSPWPFKLIRRQFPIIVSYAMTINKSQGQTLDSVG 1338
I+ +I+G ++G+ IPRM M PS S +PFK RRQ PI + +AMTINKSQG++L VG
Sbjct: 137 IQVTVITGTHIGDDISIPRMDMVPSDSSYPFKFERRQSPISLCFAMTINKSQGRSLSHVG 316
Query: 1339 LYLPRSVFSHGQLYVAFSRVRTKAGLKILIHDLEKKPLSNTTNVVYKEVFDNL 1391
LYL R V +HG LYVA RVR++ LK+L+ D E+K + T NVVY+E+F+N+
Sbjct: 317 LYLSRPVSTHG*LYVALPRVRSRK*LKLLVLDEEEKMTNTTKNVVYREIFENI 475
Score = 39.7 bits (91), Expect(2) = 5e-33
Identities = 19/26 (73%), Positives = 20/26 (76%)
Frame = +1
Query: 1246 KVGTPIMLLRNLDQVEGLCNGTRLIV 1271
K G IMLLRN+ Q GLCNGTRLIV
Sbjct: 43 KEGALIMLLRNIVQASGLCNGTRLIV 120
>BP085968
Length = 341
Score = 85.1 bits (209), Expect(2) = 9e-25
Identities = 40/73 (54%), Positives = 55/73 (74%)
Frame = +2
Query: 912 TDEQRDIYHKIMSAVNGQNGGVFFLHGYGGTGKTFMWTTLAASLRSQGKIVLTVATSGIA 971
T +Q +Y +IMS+V +GG +FL+G+GG+GKTF+W T ++ LRSQG +VL VA+SGIA
Sbjct: 11 TAKQSKVYKQIMSSVWSDDGGFYFLYGFGGSGKTFVWNTWSSGLRSQGLMVLNVASSGIA 190
Query: 972 SLLLPGGRTAHSK 984
S LLPGG+ K
Sbjct: 191 SWLLPGGKDCSFK 229
Score = 47.0 bits (110), Expect(2) = 9e-25
Identities = 23/43 (53%), Positives = 29/43 (66%)
Frame = +3
Query: 979 RTAHSKFKIPVPSFENSTCNIDGDSDLAKLLKVTNLIIWDEAP 1021
RTAHS+F I + + STCNI S A+LL+ + IIWDEAP
Sbjct: 213 RTAHSRFSIHISINDISTCNIKQGSQKAELLQKASSIIWDEAP 341
>TC8086 similar to UP|DMC1_HUMAN (Q14565) Meiotic recombination protein
DMC1/LIM15 homolog, partial (12%)
Length = 628
Score = 85.1 bits (209), Expect(2) = 3e-24
Identities = 40/75 (53%), Positives = 55/75 (73%)
Frame = +1
Query: 1257 LDQVEGLCNGTRLIVTRMANHVIEAKIISGKNVGNLTYIPRMSMSPSQSPWPFKLIRRQF 1316
+ + E LC+GTRLIV + +VI+A +I+G N+G+ +IPR+ M PS S +PFK RR F
Sbjct: 151 IQKFENLCHGTRLIVVDLGTYVIKATVITGTNIGDDIFIPRLDMVPSDSGYPFKFERR*F 330
Query: 1317 PIIVSYAMTINKSQG 1331
PI + +AMTINKSQG
Sbjct: 331 PISLCFAMTINKSQG 375
Score = 45.1 bits (105), Expect(2) = 3e-24
Identities = 20/36 (55%), Positives = 29/36 (80%)
Frame = +3
Query: 1356 SRVRTKAGLKILIHDLEKKPLSNTTNVVYKEVFDNL 1391
SRVR++ GLK+L+ D E+K + T NVVY+EVF+N+
Sbjct: 369 SRVRSRKGLKLLVLDEEEKVTNTTKNVVYREVFENI 476
>BP046554
Length = 556
Score = 74.3 bits (181), Expect(2) = 2e-17
Identities = 36/69 (52%), Positives = 45/69 (65%)
Frame = -3
Query: 372 GFPDLFITFTCNPSWPEIQRFVGAKGLKPHDRPDIIARVFKIKFDELLNDLTKKHILGKV 431
G+PDLFITFTCN +W EIQRFV + L D P+I RVFK+K D L++DL K I G
Sbjct: 254 GYPDLFITFTCNSAWCEIQRFVQPRNLNVEDCPNICVRVFKMKLDRLISDLKKGKIFGAS 75
Query: 432 VAYMYTIEF 440
A + +F
Sbjct: 74 DAVRHIHDF 48
Score = 33.1 bits (74), Expect(2) = 2e-17
Identities = 17/41 (41%), Positives = 24/41 (58%), Gaps = 8/41 (19%)
Frame = -2
Query: 335 GQTKKQG--------KRVVLPSSYVGGRRYMDQLYFDGMAI 367
G++ +QG KRV+LP S+ GG RYM + D MA+
Sbjct: 393 GRSHRQGRDESFFCWKRVILPPSFTGGHRYMFNNFKDAMAM 271
>BP065488
Length = 439
Score = 79.7 bits (195), Expect = 3e-15
Identities = 49/104 (47%), Positives = 66/104 (63%), Gaps = 2/104 (1%)
Frame = -1
Query: 1290 GNLTYIPRMSMSPSQSPWPFKLIRRQFPIIVSYAMTINKSQGQT-LDSVGLYLPRSVFSH 1348
G+ +IPRM+M PS S P K R QFPI + +AMTINKSQ +V L + +SH
Sbjct: 379 GDDIFIPRMNMVPSVSGDPLKFERCQFPISLCFAMTINKSQXSVHYPTVALISFLACYSH 200
Query: 1349 -GQLYVAFSRVRTKAGLKILIHDLEKKPLSNTTNVVYKEVFDNL 1391
+LYVA S VR++ GLK+L+ E+K + T N VY+EVF N+
Sbjct: 199 MDKLYVALSGVRSRKGLKLLVLGEEEKVTNATKNEVYQEVFKNI 68
>AU089042
Length = 191
Score = 62.4 bits (150), Expect = 5e-10
Identities = 30/59 (50%), Positives = 40/59 (66%)
Frame = +3
Query: 1245 LKVGTPIMLLRNLDQVEGLCNGTRLIVTRMANHVIEAKIISGKNVGNLTYIPRMSMSPS 1303
LK G P+ML+ NL GLCNGTRLIV + +VI A I+SG ++G + YI M++ PS
Sbjct: 9 LKXGVPVMLMXNLXISTGLCNGTRLIVDYLGPNVIGATILSGTHIGXVVYISMMNLXPS 185
>TC14212 similar to UP|Q9FN61 (Q9FN61) Gb|AAF07369.1, partial (10%)
Length = 613
Score = 41.6 bits (96), Expect = 9e-04
Identities = 22/41 (53%), Positives = 26/41 (62%)
Frame = +2
Query: 1351 LYVAFSRVRTKAGLKILIHDLEKKPLSNTTNVVYKEVFDNL 1391
LYVA SRV++K GLKILI T N+VYKEVF +
Sbjct: 2 LYVAVSRVKSKDGLKILISSDGTSTPGATKNIVYKEVFQKI 124
>TC11129 weakly similar to UP|PDA6_MEDSA (P38661) Probable protein disulfide
isomerase A6 precursor (P5) , partial (7%)
Length = 600
Score = 37.4 bits (85), Expect = 0.016
Identities = 14/40 (35%), Positives = 28/40 (70%)
Frame = +2
Query: 1182 TIDVVDKINDYVLSIIPGEEKEYFSSDSIDRSEVNDQCQS 1221
T++ V+K+N+++L ++PG EY SSD+ + + + + QS
Sbjct: 461 TLESVEKVNEFMLDLLPGNTTEYLSSDTTCKYDEDTELQS 580
>TC8119 similar to UP|Q9FFF5 (Q9FFF5) Nucleic acid binding protein-like,
partial (85%)
Length = 1249
Score = 31.2 bits (69), Expect = 1.2
Identities = 25/86 (29%), Positives = 37/86 (42%), Gaps = 10/86 (11%)
Frame = -2
Query: 942 TGKTFMWTTLAASLRSQGKIV---------LTVATSGIASLLLPGGR-TAHSKFKIPVPS 991
+G F W L+ + ++ GKI+ + V TS R T H + PVP+
Sbjct: 675 SGLVFYWLPLSDNFKNGGKIIDQAK*PFPFIAVKTSSKIKCHRKQPRITVHGNQRKPVPT 496
Query: 992 FENSTCNIDGDSDLAKLLKVTNLIIW 1017
+ ID +S L KL NL+ W
Sbjct: 495 VHSIPSKIDSESRLRKLR--WNLLCW 424
>TC7794 homologue to UP|Q9M600 (Q9M600) Chlorophyll a/b binding protein
precursor, partial (97%)
Length = 2581
Score = 30.4 bits (67), Expect = 2.0
Identities = 26/85 (30%), Positives = 38/85 (44%), Gaps = 8/85 (9%)
Frame = -1
Query: 431 VVAYMYTIEFQKRGLPHAHILIFLHP--SSKYPTP------DHINQIISAEIPHPENDRE 482
+VA T+E +K P H L+FLH S+ P DH N ++ P R+
Sbjct: 376 LVALSSTLEREK---PWLHKLLFLHHLLHSQLRLPNRALEQDHSNLLLDPPEKAPSLSRQ 206
Query: 483 LYKLVGTHMMHGPCGLAKPSSPCMK 507
L L+ + PCGL + S M+
Sbjct: 205 LLLLLSSKEQIDPCGLHQSKSLWME 131
>AV424749
Length = 416
Score = 30.4 bits (67), Expect = 2.0
Identities = 25/87 (28%), Positives = 39/87 (44%)
Frame = +2
Query: 929 QNGGVFFLHGYGGTGKTFMWTTLAASLRSQGKIVLTVATSGIASLLLPGGRTAHSKFKIP 988
Q G V +L G G+GK+ + L+ SL S+GK+ S +L G H +
Sbjct: 149 QKGCVIWLTGLSGSGKSSLACALSQSLHSRGKL----------SYVLDGDNIRHGLNRDL 298
Query: 989 VPSFENSTCNIDGDSDLAKLLKVTNLI 1015
E+ + NI ++AKL +I
Sbjct: 299 SFKAEDRSENIRRIGEVAKLFADAGVI 379
>TC8979 weakly similar to UP|Q8RY59 (Q8RY59) At1g32230/F3C3_1, partial (12%)
Length = 833
Score = 30.0 bits (66), Expect = 2.6
Identities = 16/47 (34%), Positives = 24/47 (51%)
Frame = -1
Query: 1201 EKEYFSSDSIDRSEVNDQCQSFQLFTPEFLSTLRTSGLPNHKIKLKV 1247
EK+ S ++ QC SF TP+FLS +TS +H+ K+
Sbjct: 653 EKKVPQCSSCSEKKMEYQCHSFNQITPQFLSITQTS-FQSHRCSKKL 516
>BP079996
Length = 488
Score = 28.9 bits (63), Expect = 5.8
Identities = 12/34 (35%), Positives = 20/34 (58%), Gaps = 4/34 (11%)
Frame = +2
Query: 1057 GGDFRQIL----PVVPRADRSDIINSSINSSYIW 1086
GG +R IL ++P+ R D+I+ I S++W
Sbjct: 86 GGCYRNILVAGGSIIPKGQRKDVISPPIIXSFVW 187
>BP065508
Length = 377
Score = 28.9 bits (63), Expect = 5.8
Identities = 13/36 (36%), Positives = 17/36 (47%)
Frame = +2
Query: 2 WYQERRRKSRESSNPEFGMCCGNGKIQIPFLPTPPP 37
W E+ R S+E NP G + + P P PPP
Sbjct: 38 WLMEKPRSSKEIWNPRSGSSSSSAAVMAP--PPPPP 139
>TC8167 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (65%)
Length = 2760
Score = 28.5 bits (62), Expect = 7.6
Identities = 14/30 (46%), Positives = 16/30 (52%)
Frame = -1
Query: 1221 SFQLFTPEFLSTLRTSGLPNHKIKLKVGTP 1250
S FTP F S L +S +PN K K K P
Sbjct: 2643 SIPFFTPSFQSQLCSSNIPNKKEKKKKAQP 2554
>BP063650
Length = 497
Score = 28.5 bits (62), Expect = 7.6
Identities = 13/32 (40%), Positives = 21/32 (65%)
Frame = +1
Query: 1062 QILPVVPRADRSDIINSSINSSYIWDECIVLT 1093
+ L VV + DII++ +N+SY+ D C VL+
Sbjct: 58 KFLTVVYKGRMQDIIHAIVNASYL*DHCQVLS 153
>TC14517
Length = 697
Score = 28.1 bits (61), Expect = 9.9
Identities = 25/57 (43%), Positives = 27/57 (46%), Gaps = 2/57 (3%)
Frame = +3
Query: 596 HCCSRFR-WLKSIQCK*QCR*D*TIS*-LSLCFT*RSLLEDICLSNPWKKASC*KIV 650
H C FR WL C C * IS +SL F R DICL N +ASC IV
Sbjct: 153 H*CQLFRKWLH--HCMFPCY*VQNISSHISLYFLGRDFTTDICLLNGRAEASCEAIV 317
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.336 0.147 0.471
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,514,160
Number of Sequences: 28460
Number of extensions: 328831
Number of successful extensions: 2413
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 2378
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2410
length of query: 1391
length of database: 4,897,600
effective HSP length: 102
effective length of query: 1289
effective length of database: 1,994,680
effective search space: 2571142520
effective search space used: 2571142520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148528.7


