

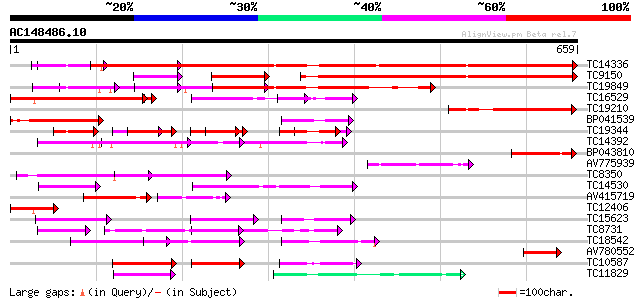
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148486.10 - phase: 0
(659 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14336 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, p... 845 0.0
TC9150 similar to UP|Q9M6E4 (Q9M6E4) Poly(A)-binding protein (Fr... 523 e-149
TC19849 similar to UP|Q9AT32 (Q9AT32) Poly(A)-binding protein, p... 272 1e-73
TC16529 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, p... 264 7e-72
TC19210 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, p... 185 2e-47
BP041539 146 1e-35
TC19344 weakly similar to GB|AAA34838.1|172092|YSCPABPG polyaden... 66 5e-22
TC14392 similar to UP|Q9LEB4 (Q9LEB4) RNA binding protein 45, pa... 94 9e-20
BP043810 83 2e-16
AV775939 77 9e-15
TC8350 similar to UP|Q9LEB4 (Q9LEB4) RNA binding protein 45, par... 72 3e-13
TC14530 similar to UP|P82277 (P82277) Plastid-specific 30S ribos... 72 4e-13
AV415719 68 5e-12
TC12406 similar to UP|Q9AT32 (Q9AT32) Poly(A)-binding protein, p... 66 2e-11
TC15623 similar to UP|P93486 (P93486) Glycine-rich RNA-binding p... 62 2e-10
TC8731 homologue to UP|Q9LNS1 (Q9LNS1) F1L3.2, partial (34%) 61 7e-10
TC18542 similar to UP|Q93YF1 (Q93YF1) Nucleic acid binding prote... 56 2e-08
AV780552 55 5e-08
TC10587 weakly similar to GB|AAC50895.1|1518802|HSU63289 CUG-BP/... 54 6e-08
TC11829 weakly similar to UP|NAM8_YEAST (Q00539) NAM8 protein, p... 53 1e-07
>TC14336 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, partial
(85%)
Length = 1917
Score = 845 bits (2182), Expect = 0.0
Identities = 423/569 (74%), Positives = 487/569 (85%), Gaps = 4/569 (0%)
Frame = +2
Query: 95 FTPMNNKSIRVMYSHRDPSSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATD 154
FTP+NN+ IR+MYSHRDPS RKSG NIFIKNLDK IDHKALHDTFS+FG I+SCK+ATD
Sbjct: 17 FTPLNNRPIRIMYSHRDPSIRKSGAGNIFIKNLDKAIDHKALHDTFSTFGNILSCKVATD 196
Query: 155 GSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFNNVY 214
SGQSKGYGFVQFE E++AQNAI+KLNGML+NDKQV+VG FLRKQ+R++ K KFNNV+
Sbjct: 197 SSGQSKGYGFVQFETEEAAQNAIEKLNGMLLNDKQVYVGPFLRKQERESSTDKAKFNNVF 376
Query: 215 VKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNGK 274
VKNLSES T+D+LKN FG +GTITSAV+MRD DG+SKCFGFVNFENA+DAA+AVE+LNGK
Sbjct: 377 VKNLSESTTDDELKNVFGEFGTITSAVVMRDGDGKSKCFGFVNFENADDAARAVESLNGK 556
Query: 275 KVDDEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKLKEM 334
KVDD+EWYVGKAQKKSEREQELK +FEQ++KE+ DK+QG NLY+KNLDDSI DEKLKE+
Sbjct: 557 KVDDKEWYVGKAQKKSEREQELKNKFEQSMKEA-ADKYQGANLYVKNLDDSIADEKLKEL 733
Query: 335 FSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKMIVS 394
FS +GTITS KV MRDPNG+SRGSGFVAFSTPEEASRAL EMNGKM+VS
Sbjct: 734 FSSYGTITSCKV------------MRDPNGISRGSGFVAFSTPEEASRALLEMNGKMVVS 877
Query: 395 KPLYVAVAQRKEDRRARLQAQFSQMRPVAITPSVAPRMPLYPPGTPGLGQQFMYGQGPPA 454
KPLYV +AQRKEDRRARLQAQF+QMRPVA+ P APR+P+YPPG PG+GQQ YGQGPPA
Sbjct: 878 KPLYVTLAQRKEDRRARLQAQFAQMRPVAMPP--APRVPMYPPGGPGIGQQIFYGQGPPA 1051
Query: 455 MMPPQAGFGYQQQLVPGMRPGGGPMPSYFVPMVQQGQQGQRPGGRRGGPGQQPPQQVPMM 514
++P Q GFGYQQQLVPGMRPGG P+P++F+PMV QGQQGQRPGGRR G QQ Q VP+M
Sbjct: 1052IIPSQPGFGYQQQLVPGMRPGGAPVPNFFMPMVPQGQQGQRPGGRRAGAVQQSQQPVPLM 1231
Query: 515 QQQMLPRQRVYRYPPGRSNIQDAPVQNIGGGMMS--YDMGGLPLRD--VVPPMPIHALAT 570
QQMLPR RVYRYPPGR + + P+ + GGM S YD+ G+PLRD + +P+ ALA+
Sbjct: 1232PQQMLPRGRVYRYPPGR-GMPEVPMPGVAGGMFSVPYDVSGMPLRDGSLSQQIPVGALAS 1408
Query: 571 ALANAPPEQQRTMLGEALYPLVDQLEHDSAAKVTGMLLEMDQPEVLHLIESPDALKAKVA 630
ALANA PEQQRTMLGE LYPLV+QLE D+AAKVTGMLLEMDQ EVLHL+ESP+ALKAKVA
Sbjct: 1409ALANASPEQQRTMLGENLYPLVEQLEPDNAAKVTGMLLEMDQTEVLHLLESPEALKAKVA 1588
Query: 631 EAMEVLRNVSQQQGNSPADQLASLSLNDS 659
EAM+VLRNV+QQQ ADQLASLSL D+
Sbjct: 1589EAMDVLRNVAQQQAGGAADQLASLSLGDN 1675
Score = 101 bits (252), Expect = 3e-22
Identities = 61/186 (32%), Positives = 102/186 (54%), Gaps = 17/186 (9%)
Frame = +2
Query: 33 SLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDV 92
+++V +L + D +L ++F + G + S V RD +S +G+VNF N DAARA++
Sbjct: 368 NVFVKNLSESTTDDELKNVFGEFGTITSAVVMRD-GDGKSKCFGFVNFENADDAARAVES 544
Query: 93 LNFTPMNNKSIRV-----------------MYSHRDPSSRKSGTANIFIKNLDKTIDHKA 135
LN +++K V S ++ + + G AN+++KNLD +I +
Sbjct: 545 LNGKKVDDKEWYVGKAQKKSEREQELKNKFEQSMKEAADKYQG-ANLYVKNLDDSIADEK 721
Query: 136 LHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHF 195
L + FSS+G I SCK+ D +G S+G GFV F + A A+ ++NG ++ K ++V
Sbjct: 722 LKELFSSYGTITSCKVMRDPNGISRGSGFVAFSTPEEASRALLEMNGKMVVSKPLYVTLA 901
Query: 196 LRKQDR 201
RK+DR
Sbjct: 902 QRKEDR 919
Score = 49.7 bits (117), Expect = 2e-06
Identities = 29/90 (32%), Positives = 52/90 (57%)
Frame = +2
Query: 26 ANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQD 85
A+++ +LYV +L+ ++ D +L +LF+ G + S +V RD S G G+V F+ P++
Sbjct: 656 ADKYQGANLYVKNLDDSIADEKLKELFSSYGTITSCKVMRD-PNGISRGSGFVAFSTPEE 832
Query: 86 AARALDVLNFTPMNNKSIRVMYSHRDPSSR 115
A+RAL +N + +K + V + R R
Sbjct: 833 ASRALLEMNGKMVVSKPLYVTLAQRKEDRR 922
>TC9150 similar to UP|Q9M6E4 (Q9M6E4) Poly(A)-binding protein (Fragment),
partial (65%)
Length = 1191
Score = 523 bits (1347), Expect = e-149
Identities = 267/324 (82%), Positives = 287/324 (88%), Gaps = 3/324 (0%)
Frame = +3
Query: 339 GTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKMIVSKPLY 398
GTITS+K IMRDP G+SRGSGFVAFSTPEEASRALGEMNGKMI KPLY
Sbjct: 27 GTITSFK------------IMRDPTGISRGSGFVAFSTPEEASRALGEMNGKMISGKPLY 170
Query: 399 VAVAQRKEDRRARLQAQFSQMRPVAITPSVAPRMPLYPPGTPGLGQQFMYGQGPPAMMPP 458
VA+AQRKEDRRARLQAQFSQMRPVAI+PSVAPRMP+YPPG PGLGQQ++YGQGPPAM+P
Sbjct: 171 VALAQRKEDRRARLQAQFSQMRPVAISPSVAPRMPIYPPGAPGLGQQYLYGQGPPAMIPQ 350
Query: 459 QAGFGYQQQLVPGMRPGGGPMPSYFVPMVQQGQQGQRPGGRRGGPGQQPPQQVPMMQQQM 518
QAGFGYQQQL+PGMRPGG PM S+FVPMVQQGQQGQRPGGRR G QQP Q +PMMQQQM
Sbjct: 351 QAGFGYQQQLMPGMRPGGAPMQSFFVPMVQQGQQGQRPGGRRAGHVQQPQQPMPMMQQQM 530
Query: 519 LPRQRVYRYPPGRSNIQDAPVQNIGGGMMS--YDMGGLPLRDVV-PPMPIHALATALANA 575
LPR RVYRYPPGR N+QD P+Q + GGMMS YDMGGLP+RD V PMPI ALATALANA
Sbjct: 531 LPRGRVYRYPPGR-NMQDGPMQGVAGGMMSVPYDMGGLPIRDPVGQPMPIQALATALANA 707
Query: 576 PPEQQRTMLGEALYPLVDQLEHDSAAKVTGMLLEMDQPEVLHLIESPDALKAKVAEAMEV 635
P EQQRTMLGEALYPLVDQLEHDSAAKVTGMLLEMDQPEVLHLIESPDALKAKVAEA++V
Sbjct: 708 PLEQQRTMLGEALYPLVDQLEHDSAAKVTGMLLEMDQPEVLHLIESPDALKAKVAEALDV 887
Query: 636 LRNVSQQQGNSPADQLASLSLNDS 659
LRNV+QQQ N+PADQLASLSLND+
Sbjct: 888 LRNVAQQQANNPADQLASLSLNDN 959
Score = 58.9 bits (141), Expect = 2e-09
Identities = 29/68 (42%), Positives = 42/68 (61%)
Frame = +3
Query: 235 GTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQKKSEREQ 294
GTITS +MRD G S+ GFV F E+A++A+ +NGK + + YV AQ+K +R
Sbjct: 27 GTITSFKIMRDPTGISRGSGFVAFSTPEEASRALGEMNGKMISGKPLYVALAQRKEDRRA 206
Query: 295 ELKGRFEQ 302
L+ +F Q
Sbjct: 207 RLQAQFSQ 230
Score = 48.5 bits (114), Expect = 3e-06
Identities = 24/58 (41%), Positives = 35/58 (59%)
Frame = +3
Query: 144 GQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDR 201
G I S KI D +G S+G GFV F + A A+ ++NG +I+ K ++V RK+DR
Sbjct: 27 GTITSFKIMRDPTGISRGSGFVAFSTPEEASRALGEMNGKMISGKPLYVALAQRKEDR 200
Score = 37.4 bits (85), Expect = 0.008
Identities = 21/61 (34%), Positives = 34/61 (55%)
Frame = +3
Query: 55 VGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDVLNFTPMNNKSIRVMYSHRDPSS 114
VG + S ++ RD T S G G+V F+ P++A+RAL +N ++ K + V + R
Sbjct: 24 VGTITSFKIMRD-PTGISRGSGFVAFSTPEEASRALGEMNGKMISGKPLYVALAQRKEDR 200
Query: 115 R 115
R
Sbjct: 201 R 203
>TC19849 similar to UP|Q9AT32 (Q9AT32) Poly(A)-binding protein, partial
(30%)
Length = 603
Score = 272 bits (695), Expect = 1e-73
Identities = 149/260 (57%), Positives = 180/260 (68%)
Frame = +2
Query: 236 TITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQKKSEREQE 295
TITSAV+MRD DG+SKCFGFVNFENA++AAKAV+ALNGKK DD+EWYVGKA+KKSERE E
Sbjct: 2 TITSAVVMRDVDGKSKCFGFVNFENADEAAKAVDALNGKKFDDKEWYVGKAKKKSERELE 181
Query: 296 LKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDI 355
LK + EQ+++E+ VDK+QG NLYLKNLDD++ DEKL+E+FSEFGTITS K
Sbjct: 182 LKEQHEQSMQET-VDKYQGANLYLKNLDDNVDDEKLRELFSEFGTITSCK---------- 328
Query: 356 RVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRKEDRRARLQAQ 415
IMRDP+GVSRGSGFVAFSTPEEA+RALGEMNGKM+ KPLYVA+AQ+KE+RRA+LQ
Sbjct: 329 --IMRDPHGVSRGSGFVAFSTPEEATRALGEMNGKMVDGKPLYVALAQKKEERRAKLQGG 502
Query: 416 FSQMRPVAITPSVAPRMPLYPPGTPGLGQQFMYGQGPPAMMPPQAGFGYQQQLVPGMRPG 475
+ Q QQ + G P G
Sbjct: 503 YFQ-------------------------QQLLPGMRP----------------------G 541
Query: 476 GGPMPSYFVPMVQQGQQGQR 495
GGP+ +++VP+ QGQQGQR
Sbjct: 542 GGPLSNFYVPLGHQGQQGQR 601
Score = 117 bits (293), Expect = 6e-27
Identities = 65/169 (38%), Positives = 95/169 (55%), Gaps = 12/169 (7%)
Frame = +2
Query: 146 IMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDRD--- 202
I S + D G+SK +GFV FE D A A+D LNG +DK+ +VG +K +R+
Sbjct: 5 ITSAVVMRDVDGKSKCFGFVNFENADEAAKAVDALNGKKFDDKEWYVGKAKKKSERELEL 184
Query: 203 ---------NVLSKTKFNNVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCF 253
+ K + N+Y+KNL ++ ++ L+ F +GTITS +MRD G S+
Sbjct: 185 KEQHEQSMQETVDKYQGANLYLKNLDDNVDDEKLRELFSEFGTITSCKIMRDPHGVSRGS 364
Query: 254 GFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQKKSEREQELKGRFEQ 302
GFV F E+A +A+ +NGK VD + YV AQKK ER +L+G + Q
Sbjct: 365 GFVAFSTPEEATRALGEMNGKMVDGKPLYVALAQKKEERRAKLQGGYFQ 511
Score = 89.4 bits (220), Expect = 2e-18
Identities = 52/160 (32%), Positives = 83/160 (51%), Gaps = 16/160 (10%)
Frame = +2
Query: 58 VVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDVLNFTPMNNKS-------------IR 104
+ S V RD+ +S +G+VNF N +AA+A+D LN ++K +
Sbjct: 5 ITSAVVMRDV-DGKSKCFGFVNFENADEAAKAVDALNGKKFDDKEWYVGKAKKKSERELE 181
Query: 105 VMYSHRDPSSR---KSGTANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKG 161
+ H K AN+++KNLD +D + L + FS FG I SCKI D G S+G
Sbjct: 182 LKEQHEQSMQETVDKYQGANLYLKNLDDNVDDEKLRELFSEFGTITSCKIMRDPHGVSRG 361
Query: 162 YGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDR 201
GFV F + A A+ ++NG +++ K ++V +K++R
Sbjct: 362 SGFVAFSTPEEATRALGEMNGKMVDGKPLYVALAQKKEER 481
Score = 51.2 bits (121), Expect = 5e-07
Identities = 29/101 (28%), Positives = 55/101 (53%)
Frame = +2
Query: 27 NQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDA 86
+++ +LY+ +L+ NV+D +L +LF++ G + S ++ RD S G G+V F+ P++A
Sbjct: 221 DKYQGANLYLKNLDDNVDDEKLRELFSEFGTITSCKIMRD-PHGVSRGSGFVAFSTPEEA 397
Query: 87 ARALDVLNFTPMNNKSIRVMYSHRDPSSRKSGTANIFIKNL 127
RAL +N ++ K + V + + R F + L
Sbjct: 398 TRALGEMNGKMVDGKPLYVALAQKKEERRAKLQGGYFQQQL 520
>TC16529 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, partial
(22%)
Length = 761
Score = 264 bits (675), Expect(2) = 7e-72
Identities = 137/166 (82%), Positives = 142/166 (85%), Gaps = 6/166 (3%)
Frame = +1
Query: 1 MAQIQVQHQTP--APVPAPSNGVVPNVAN----QFVTTSLYVGDLEVNVNDSQLYDLFNQ 54
MAQIQVQHQTP A PA NG VA FVTTSLYVGDL+ NVNDSQLYDLFNQ
Sbjct: 220 MAQIQVQHQTPPVAAGPATPNGAAGGVAPAPNANFVTTSLYVGDLDQNVNDSQLYDLFNQ 399
Query: 55 VGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDVLNFTPMNNKSIRVMYSHRDPSS 114
VGQVVSVRVCRDL TRRSLGYGYVNF+NP DAARALDVLNFTP+NN+ IRVMYSHRDPS
Sbjct: 400 VGQVVSVRVCRDLTTRRSLGYGYVNFSNPPDAARALDVLNFTPLNNRPIRVMYSHRDPSI 579
Query: 115 RKSGTANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSK 160
RKSGTANIFIKNLDK IDHKALHDTFSSFG I+SCKIATD SGQSK
Sbjct: 580 RKSGTANIFIKNLDKAIDHKALHDTFSSFGSILSCKIATDASGQSK 717
Score = 60.5 bits (145), Expect = 9e-10
Identities = 41/138 (29%), Positives = 69/138 (49%), Gaps = 1/138 (0%)
Frame = +1
Query: 212 NVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRD-ADGRSKCFGFVNFENAEDAAKAVEA 270
++YV +L ++ + L + F G + S + RD RS +G+VNF N DAA+A++
Sbjct: 334 SLYVGDLDQNVNDSQLYDLFNQVGQVVSVRVCRDLTTRRSLGYGYVNFSNPPDAARALDV 513
Query: 271 LNGKKVDDEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEK 330
LN +++ V S R+ ++ K N+++KNLD +I +
Sbjct: 514 LNFTPLNNRPIRV----MYSHRDPSIR-------------KSGTANIFIKNLDKAIDHKA 642
Query: 331 LKEMFSEFGTITSYKVCT 348
L + FS FG+I S K+ T
Sbjct: 643 LHDTFSSFGSILSCKIAT 696
Score = 48.5 bits (114), Expect = 3e-06
Identities = 29/93 (31%), Positives = 51/93 (54%)
Frame = +1
Query: 312 FQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGF 371
F +LY+ +LD ++ D +L ++F++ G + S +VC + T R S G G+
Sbjct: 322 FVTTSLYVGDLDQNVNDSQLYDLFNQVGQVVSVRVCRDL--TTRR---------SLGYGY 468
Query: 372 VAFSTPEEASRALGEMNGKMIVSKPLYVAVAQR 404
V FS P +A+RAL +N + ++P+ V + R
Sbjct: 469 VNFSNPPDAARALDVLNFTPLNNRPIRVMYSHR 567
Score = 24.3 bits (51), Expect(2) = 7e-72
Identities = 9/14 (64%), Positives = 11/14 (78%)
Frame = +3
Query: 157 GQSKGYGFVQFEAE 170
G +GYGFVQF+ E
Sbjct: 705 GPVQGYGFVQFDTE 746
>TC19210 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, partial
(19%)
Length = 560
Score = 185 bits (469), Expect = 2e-47
Identities = 100/148 (67%), Positives = 115/148 (77%)
Frame = +1
Query: 511 VPMMQQQMLPRQRVYRYPPGRSNIQDAPVQNIGGGMMSYDMGGLPLRDVVPPMPIHALAT 570
VP+MQQQM PR RVYRY PG N+Q+ P+ GG+ + I ALAT
Sbjct: 1 VPLMQQQMHPRGRVYRYRPGH-NLQNVPLPGAAGGV---------------GVSIQALAT 132
Query: 571 ALANAPPEQQRTMLGEALYPLVDQLEHDSAAKVTGMLLEMDQPEVLHLIESPDALKAKVA 630
ALA+A PE+QRTMLGE LYPLVD+LEHD+AAK+TGMLLEMDQ EVLHLIESPDALKAKVA
Sbjct: 133 ALADATPEKQRTMLGEVLYPLVDKLEHDAAAKLTGMLLEMDQTEVLHLIESPDALKAKVA 312
Query: 631 EAMEVLRNVSQQQGNSPADQLASLSLND 658
EAM+VLRNV++QQ N+P DQLASLSLND
Sbjct: 313 EAMDVLRNVARQQSNTPIDQLASLSLND 396
>BP041539
Length = 521
Score = 146 bits (368), Expect = 1e-35
Identities = 76/109 (69%), Positives = 86/109 (78%)
Frame = +2
Query: 1 MAQIQVQHQTPAPVPAPSNGVVPNVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVS 60
MAQI H+T A V ANQ + TSLYVGDL+ +NDSQLYDLFNQ+GQVVS
Sbjct: 230 MAQI---HETDA---------VAQEANQPMPTSLYVGDLDTEINDSQLYDLFNQIGQVVS 373
Query: 61 VRVCRDLATRRSLGYGYVNFTNPQDAARALDVLNFTPMNNKSIRVMYSH 109
VRVCRDLAT++SLGYGYVNFTNP+DAA ALDVLNFTP+N K IR+MYSH
Sbjct: 374 VRVCRDLATQQSLGYGYVNFTNPKDAATALDVLNFTPLNGKPIRIMYSH 520
Score = 44.7 bits (104), Expect = 5e-05
Identities = 24/84 (28%), Positives = 44/84 (51%)
Frame = +2
Query: 316 NLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFS 375
+LY+ +LD I D +L ++F++ G + S +VC + S G G+V F+
Sbjct: 290 SLYVGDLDTEINDSQLYDLFNQIGQVVSVRVCRDLATQQ-----------SLGYGYVNFT 436
Query: 376 TPEEASRALGEMNGKMIVSKPLYV 399
P++A+ AL +N + KP+ +
Sbjct: 437 NPKDAATALDVLNFTPLNGKPIRI 508
Score = 40.4 bits (93), Expect = 0.001
Identities = 22/62 (35%), Positives = 35/62 (55%), Gaps = 1/62 (1%)
Frame = +2
Query: 212 NVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRD-ADGRSKCFGFVNFENAEDAAKAVEA 270
++YV +L + L + F G + S + RD A +S +G+VNF N +DAA A++
Sbjct: 290 SLYVGDLDTEINDSQLYDLFNQIGQVVSVRVCRDLATQQSLGYGYVNFTNPKDAATALDV 469
Query: 271 LN 272
LN
Sbjct: 470 LN 475
>TC19344 weakly similar to GB|AAA34838.1|172092|YSCPABPG
polyadenylate-binding protein {Saccharomyces
cerevisiae;} , partial (18%)
Length = 518
Score = 65.9 bits (159), Expect(2) = 5e-22
Identities = 33/71 (46%), Positives = 48/71 (67%)
Frame = +3
Query: 314 GLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVA 373
G N+Y+KN+DD+++DE+L+ FS G+IT KV M+D G+S+G GFV
Sbjct: 150 GSNIYVKNIDDNVSDEELRGHFSACGSITYAKV------------MKDEKGISQGFGFVC 293
Query: 374 FSTPEEASRAL 384
FSTPEEA++A+
Sbjct: 294 FSTPEEANKAV 326
Score = 57.0 bits (136), Expect(2) = 1e-18
Identities = 26/58 (44%), Positives = 41/58 (69%)
Frame = +3
Query: 211 NNVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAV 268
+N+YVKN+ ++ ++++L+ F A G+IT A +M+D G S+ FGFV F E+A KAV
Sbjct: 153 SNIYVKNIDDNVSDEELRGHFSACGSITYAKVMKDEKGISQGFGFVCFSTPEEANKAV 326
Score = 55.8 bits (133), Expect(2) = 5e-22
Identities = 23/49 (46%), Positives = 37/49 (74%)
Frame = +2
Query: 228 KNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNGKKV 276
K +F ++G I S + +D +G SK FGFVN+EN++DA +A+EA+NG ++
Sbjct: 2 KEKFSSFGKIVSLTISKDENGMSKGFGFVNYENSDDAKRALEAMNGTQL 148
Score = 53.1 bits (126), Expect(2) = 1e-18
Identities = 24/57 (42%), Positives = 37/57 (64%)
Frame = +2
Query: 138 DTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGH 194
+ FSSFG+I+S I+ D +G SKG+GFV +E D A+ A++ +NG + K + H
Sbjct: 5 EKFSSFGKIVSLTISKDENGMSKGFGFVNYENSDDAKRALEAMNGTQLGIKHLCEEH 175
Score = 48.1 bits (113), Expect(2) = 3e-12
Identities = 21/59 (35%), Positives = 34/59 (57%)
Frame = +3
Query: 120 ANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAID 178
+NI++KN+D + + L FS+ G I K+ D G S+G+GFV F + A A++
Sbjct: 153 SNIYVKNIDDNVSDEELRGHFSACGSITYAKVMKDEKGISQGFGFVCFSTPEEANKAVN 329
Score = 43.5 bits (101), Expect = 1e-04
Identities = 26/66 (39%), Positives = 33/66 (49%)
Frame = +2
Query: 332 KEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKM 391
KE FS FG I S I +D NG+S+G GFV + ++A RAL MNG
Sbjct: 2 KEKFSSFGKIVSL------------TISKDENGMSKGFGFVNYENSDDAKRALEAMNGTQ 145
Query: 392 IVSKPL 397
+ K L
Sbjct: 146 LGIKHL 163
Score = 40.4 bits (93), Expect(2) = 3e-12
Identities = 19/52 (36%), Positives = 32/52 (61%)
Frame = +2
Query: 52 FNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDVLNFTPMNNKSI 103
F+ G++VS+ + +D S G+G+VN+ N DA RAL+ +N T + K +
Sbjct: 11 FSSFGKIVSLTISKD-ENGMSKGFGFVNYENSDDAKRALEAMNGTQLGIKHL 163
>TC14392 similar to UP|Q9LEB4 (Q9LEB4) RNA binding protein 45, partial (77%)
Length = 1576
Score = 93.6 bits (231), Expect = 9e-20
Identities = 69/265 (26%), Positives = 127/265 (47%), Gaps = 24/265 (9%)
Frame = +3
Query: 33 SLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDV 92
+L++GDL+ ++++ LY F G++ +V+V R+ T +S GYG++ FT+ A R L
Sbjct: 336 TLWIGDLQYWMDENYLYQCFAHTGELATVKVIRNKNTNQSEGYGFLEFTSRAGAERILQT 515
Query: 93 LNFTPMNN--KSIRVMYSHRDPSSRK----SGTANIFIKNLDKTIDHKALHDTF-SSFGQ 145
N T M N ++ R+ ++ + S IF+ +L + L + F + +
Sbjct: 516 YNGTIMPNGGQNFRLNWATFSAGGERRHDDSPDYTIFVGDLAADVTDYHLTEVFRTRYNS 695
Query: 146 IMSCKIATDG-SGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRK------ 198
+ K+ D + ++KGYGFV+F E A+ ++ G++ + + + +G K
Sbjct: 696 VKGAKVVIDRLTSRTKGYGFVRFADESEQVRAMTEMQGVVCSTRPMRIGPAANKNLGTGT 875
Query: 199 --------QDRDNVLSKTKFNN--VYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADG 248
Q+ ++ NN ++V NL + T+D L+ FG YG + +
Sbjct: 876 GTQPKASYQNSQGPQNENDPNNTTIFVGNLDPNVTDDHLRQVFGLYGDLVHVKI-----P 1040
Query: 249 RSKCFGFVNFENAEDAAKAVEALNG 273
+ K GFV F + A +A+ LNG
Sbjct: 1041QGKRCGFVQFADRSCAEEALRVLNG 1115
Score = 71.6 bits (174), Expect = 4e-13
Identities = 66/301 (21%), Positives = 129/301 (41%), Gaps = 15/301 (4%)
Frame = +3
Query: 107 YSHRDPSSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATD-GSGQSKGYGFV 165
+ H PS+ ++I +L +D L+ F+ G++ + K+ + + QS+GYGF+
Sbjct: 297 HQHAQPSTPDE-VRTLWIGDLQYWMDENYLYQCFAHTGELATVKVIRNKNTNQSEGYGFL 473
Query: 166 QFEAEDSAQNAIDKLNGMLI-NDKQVF--------VGHFLRKQDRDNVLSKTKFNNVYVK 216
+F + A+ + NG ++ N Q F G R D + ++V
Sbjct: 474 EFTSRAGAERILQTYNGTIMPNGGQNFRLNWATFSAGGERRHDDSPDY-------TIFVG 632
Query: 217 NLSESFTEDDLKNEFGA-YGTITSAVLMRDA-DGRSKCFGFVNFENAEDAAKAVEALNGK 274
+L+ T+ L F Y ++ A ++ D R+K +GFV F + + +A+ + G
Sbjct: 633 DLAADVTDYHLTEVFRTRYNSVKGAKVVIDRLTSRTKGYGFVRFADESEQVRAMTEMQGV 812
Query: 275 KVDDEEWYVGKAQKKS---EREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKL 331
+G A K+ + K ++ + + +++ NLD ++TD+ L
Sbjct: 813 VCSTRPMRIGPAANKNLGTGTGTQPKASYQNSQGPQNENDPNNTTIFVGNLDPNVTDDHL 992
Query: 332 KEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKM 391
+++F +G + K+ P G + GFV F+ A AL +NG +
Sbjct: 993 RQVFGLYGDLVHVKI---------------PQG--KRCGFVQFADRSCAEEALRVLNGTL 1121
Query: 392 I 392
+
Sbjct: 1122L 1124
Score = 66.6 bits (161), Expect = 1e-11
Identities = 51/201 (25%), Positives = 86/201 (42%), Gaps = 21/201 (10%)
Frame = +3
Query: 33 SLYVGDLEVNVNDSQLYDLFN-QVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALD 91
+++VGDL +V D L ++F + V +V D T R+ GYG+V F + + RA+
Sbjct: 618 TIFVGDLAADVTDYHLTEVFRTRYNSVKGAKVVIDRLTSRTKGYGFVRFADESEQVRAMT 797
Query: 92 VLN----------FTPMNNKSI--------RVMYSHRDPSSRKSGTAN--IFIKNLDKTI 131
+ P NK++ + Y + ++ N IF+ NLD +
Sbjct: 798 EMQGVVCSTRPMRIGPAANKNLGTGTGTQPKASYQNSQGPQNENDPNNTTIFVGNLDPNV 977
Query: 132 DHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVF 191
L F +G ++ KI Q K GFVQF A+ A+ LNG L+ + V
Sbjct: 978 TDDHLRQVFGLYGDLVHVKIP-----QGKRCGFVQFADRSCAEEALRVLNGTLLGGQNVR 1142
Query: 192 VGHFLRKQDRDNVLSKTKFNN 212
+ ++ +++NN
Sbjct: 1143LSWGRSPSNKQTQSDPSQWNN 1205
Score = 28.9 bits (63), Expect = 2.7
Identities = 16/37 (43%), Positives = 19/37 (51%), Gaps = 2/37 (5%)
Frame = +3
Query: 495 RPGGRRGGPGQQPPQQVPMMQQ--QMLPRQRVYRYPP 529
+PGG P QQP QQ QQ M+P Q+ PP
Sbjct: 126 QPGGPGMAPPQQPNQQYQQQQQPYMMMPPQQHQPQPP 236
>BP043810
Length = 500
Score = 82.8 bits (203), Expect = 2e-16
Identities = 46/76 (60%), Positives = 57/76 (74%), Gaps = 1/76 (1%)
Frame = -3
Query: 584 LGEALYPLVDQLE-HDSAAKVTGMLLEMDQPEVLHLIESPDALKAKVAEAMEVLRNVSQQ 642
LGE LYPLV+ L + AKVTGMLLEMDQ EV+HLIESP+ LK KV+EA+ VLR+ +
Sbjct: 498 LGEHLYPLVEHLTPNQHTAKVTGMLLEMDQSEVIHLIESPEDLKMKVSEALRVLRDAA-A 322
Query: 643 QGNSPADQLASLSLND 658
G+ +DQL S SLN+
Sbjct: 321 PGSEVSDQLGSFSLNE 274
>AV775939
Length = 409
Score = 77.0 bits (188), Expect = 9e-15
Identities = 54/129 (41%), Positives = 72/129 (54%), Gaps = 5/129 (3%)
Frame = +1
Query: 416 FSQMRPVAITPSVAPRMPLYPPGTPGLGQQ-FMYGQGPPAMMPPQA-GFGYQQQLVPGMR 473
F+Q++ + P A +P Y GTP L Q YGQG P MPPQ G+G+QQQ++PGMR
Sbjct: 13 FAQIKSGGMVPLPAG-IPAYHAGTPRLAPQ*LYYGQGSPGFMPPQPDGYGFQQQMLPGMR 189
Query: 474 PGGGPMPSYFVP--MVQQGQQGQRPGGRRGGPGQQ-PPQQVPMMQQQMLPRQRVYRYPPG 530
G P++ +P + +QGQ GQR GGR G Q P+QVP R RVY
Sbjct: 190 --AGVAPNFIMPYQVQRQGQPGQRVGGRGNGNLQSGQPKQVPHRNS----RARVY-ISGH 348
Query: 531 RSNIQDAPV 539
+SN+ P+
Sbjct: 349 QSNLAPGPI 375
>TC8350 similar to UP|Q9LEB4 (Q9LEB4) RNA binding protein 45, partial (38%)
Length = 735
Score = 72.0 bits (175), Expect = 3e-13
Identities = 47/166 (28%), Positives = 82/166 (49%), Gaps = 7/166 (4%)
Frame = +1
Query: 9 QTPAPVPAPSNGVVPNVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLA 68
QTP PA + V +L++GDL+ ++++ LY F G+V +V+V R+
Sbjct: 259 QTPQQQPATAEEV----------RTLWIGDLQYWMDENYLYTCFAHTGEVTTVKVIRNKQ 408
Query: 69 TRRSLGYGYVNFTNPQDAARALDVLN--FTPMNNKSIRVMYSHRDPSSRKSGTA---NIF 123
T +S GYG++ F + A R L N P ++ R+ ++ ++ IF
Sbjct: 409 TSQSEGYGFIEFNSRAAAERVLQTFNGAIMPNGGQNFRLNWASLSAGEKRHDDTPDYTIF 588
Query: 124 IKNLDKTIDHKALHDTF-SSFGQIMSCKIATDG-SGQSKGYGFVQF 167
+ +L + L +TF + + + K+ D SG++KGYGFV+F
Sbjct: 589 VGDLAADVTDYLLQETFRARYNSVKGAKVVIDRLSGRTKGYGFVRF 726
Score = 52.8 bits (125), Expect = 2e-07
Identities = 37/141 (26%), Positives = 73/141 (51%), Gaps = 4/141 (2%)
Frame = +1
Query: 122 IFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDG-SGQSKGYGFVQFEAEDSAQNAIDKL 180
++I +L +D L+ F+ G++ + K+ + + QS+GYGF++F + +A+ +
Sbjct: 304 LWIGDLQYWMDENYLYTCFAHTGEVTTVKVIRNKQTSQSEGYGFIEFNSRAAAERVLQTF 483
Query: 181 NGMLI-NDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEFGA-YGTIT 238
NG ++ N Q F ++ + T ++V +L+ T+ L+ F A Y ++
Sbjct: 484 NGAIMPNGGQNFRLNWASLSAGEKRHDDTPDYTIFVGDLAADVTDYLLQETFRARYNSVK 663
Query: 239 SAVLMRD-ADGRSKCFGFVNF 258
A ++ D GR+K +GFV F
Sbjct: 664 GAKVVIDRLSGRTKGYGFVRF 726
Score = 30.4 bits (67), Expect = 0.95
Identities = 25/103 (24%), Positives = 36/103 (34%)
Frame = +1
Query: 416 FSQMRPVAITPSVAPRMPLYPPGTPGLGQQFMYGQGPPAMMPPQAGFGYQQQLVPGMRPG 475
FS ++++PS + PG +G PP M+ Q YQQ
Sbjct: 43 FSLYPSLSLSPSRFSVREMMQPG---------HGMAPPTMVQQQPAQQYQQP-------- 171
Query: 476 GGPMPSYFVPMVQQGQQGQRPGGRRGGPGQQPPQQVPMMQQQM 518
P+ P Q Q P P Q PQQ P +++
Sbjct: 172 -PPLQPQQPPYAMMPPQAQAPPPHMWAPNVQTPQQQPATAEEV 297
>TC14530 similar to UP|P82277 (P82277) Plastid-specific 30S ribosomal
protein 2, chloroplast precursor (PSRP-2), partial (68%)
Length = 1104
Score = 71.6 bits (174), Expect = 4e-13
Identities = 56/193 (29%), Positives = 84/193 (43%), Gaps = 1/193 (0%)
Frame = +2
Query: 213 VYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDA-DGRSKCFGFVNFENAEDAAKAVEAL 271
+YV N+ + T D+L N + + A +M D GRS+ F FV + EDA A+E L
Sbjct: 236 LYVGNIPRTLTNDELANIVQEHAAVEKAEVMYDKYSGRSRRFAFVTVKTVEDANAAIEKL 415
Query: 272 NGKKVDDEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKL 331
NG ++ E V +K + Q + VD +Y+ NL ++T + L
Sbjct: 416 NGTQIGGREIKVNVTEKPL---TTVDFPLVQAEESEYVD--SPYKVYVGNLAKTVTSDML 580
Query: 332 KEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKM 391
K FSE G + S KV + S G GFV FS+ E+ A+ N
Sbjct: 581 KNFFSEKGKVLSAKVSRV-----------PGTSKSSGFGFVTFSSDEDVEAAISSFNNAP 727
Query: 392 IVSKPLYVAVAQR 404
+ + + V A R
Sbjct: 728 LEGQKIRVNKA*R 766
Score = 53.5 bits (127), Expect = 1e-07
Identities = 25/72 (34%), Positives = 42/72 (57%)
Frame = +2
Query: 34 LYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDVL 93
+YVG+L V L + F++ G+V+S +V R T +S G+G+V F++ +D A+
Sbjct: 536 VYVGNLAKTVTSDMLKNFFSEKGKVLSAKVSRVPGTSKSSGFGFVTFSSDEDVEAAISSF 715
Query: 94 NFTPMNNKSIRV 105
N P+ + IRV
Sbjct: 716 NNAPLEGQKIRV 751
>AV415719
Length = 426
Score = 67.8 bits (164), Expect = 5e-12
Identities = 32/80 (40%), Positives = 50/80 (62%)
Frame = +3
Query: 86 AARALDVLNFTPMNNKSIRVMYSHRDPSSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQ 145
A +A++V N + +N K IRV++S RDP RK+ N+F+KNL +++D L + F +G
Sbjct: 195 AMQAIEVKNHSTLNGKVIRVLWSRRDPDIRKNCIGNVFVKNLAESVDSAGLEEIFKKYGN 374
Query: 146 IMSCKIATDGSGQSKGYGFV 165
I+ + G+ KGYGFV
Sbjct: 375 IL---VVMADDGKIKGYGFV 425
Score = 42.4 bits (98), Expect = 2e-04
Identities = 33/85 (38%), Positives = 43/85 (49%)
Frame = +3
Query: 172 SAQNAIDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEF 231
SA AI+ N +N K + V R D + K NV+VKNL+ES L+ F
Sbjct: 192 SAMQAIEVKNHSTLNGKVIRVLWSRRDPD----IRKNCIGNVFVKNLAESVDSAGLEEIF 359
Query: 232 GAYGTITSAVLMRDADGRSKCFGFV 256
YG I V+M D DG+ K +GFV
Sbjct: 360 KKYGNI--LVVMAD-DGKIKGYGFV 425
>TC12406 similar to UP|Q9AT32 (Q9AT32) Poly(A)-binding protein, partial (6%)
Length = 450
Score = 66.2 bits (160), Expect = 2e-11
Identities = 37/61 (60%), Positives = 40/61 (64%), Gaps = 5/61 (8%)
Frame = +2
Query: 1 MAQIQVQHQTPAPVPAPSNGVVPNVA-----NQFVTTSLYVGDLEVNVNDSQLYDLFNQV 55
MAQ+QVQ Q P P G A NQFVTTSLYVGDL+ NV DSQLYDLFNQ+
Sbjct: 266 MAQVQVQPQNAMPGPNGGGGGGAAAAGGAGNNQFVTTSLYVGDLDPNVTDSQLYDLFNQL 445
Query: 56 G 56
G
Sbjct: 446 G 448
>TC15623 similar to UP|P93486 (P93486) Glycine-rich RNA-binding protein
PsGRBP, partial (88%)
Length = 683
Score = 62.4 bits (150), Expect = 2e-10
Identities = 30/88 (34%), Positives = 51/88 (57%)
Frame = +1
Query: 31 TTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARAL 90
++ L++G L V+D L D F+ G V RV D T RS G+G+V+F + + A+ AL
Sbjct: 181 SSKLFIGGLSYGVDDKSLEDAFSSFGTVAEARVIVDRDTGRSRGFGFVSFDSEESASSAL 360
Query: 91 DVLNFTPMNNKSIRVMYSHRDPSSRKSG 118
++ +N ++IRV Y++ P+ + G
Sbjct: 361 SSMDGQDLNGRNIRVSYANDRPAGPRGG 444
Score = 49.7 bits (117), Expect = 2e-06
Identities = 29/86 (33%), Positives = 46/86 (52%)
Frame = +1
Query: 317 LYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFST 376
L++ L + D+ L++ FS FGT+ + RVI+ G SRG GFV+F +
Sbjct: 190 LFIGGLSYGVDDKSLEDAFSSFGTVA-----------EARVIVDRDTGRSRGFGFVSFDS 336
Query: 377 PEEASRALGEMNGKMIVSKPLYVAVA 402
E AS AL M+G+ + + + V+ A
Sbjct: 337 EESASSALSSMDGQDLNGRNIRVSYA 414
Score = 47.8 bits (112), Expect = 6e-06
Identities = 23/80 (28%), Positives = 48/80 (59%), Gaps = 1/80 (1%)
Frame = +1
Query: 211 NNVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDAD-GRSKCFGFVNFENAEDAAKAVE 269
+ +++ LS + L++ F ++GT+ A ++ D D GRS+ FGFV+F++ E A+ A+
Sbjct: 184 SKLFIGGLSYGVDDKSLEDAFSSFGTVAEARVIVDRDTGRSRGFGFVSFDSEESASSALS 363
Query: 270 ALNGKKVDDEEWYVGKAQKK 289
+++G+ ++ V A +
Sbjct: 364 SMDGQDLNGRNIRVSYANDR 423
>TC8731 homologue to UP|Q9LNS1 (Q9LNS1) F1L3.2, partial (34%)
Length = 721
Score = 60.8 bits (146), Expect = 7e-10
Identities = 45/162 (27%), Positives = 77/162 (46%), Gaps = 1/162 (0%)
Frame = +2
Query: 111 DPSSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAE 170
DPS+ +S +++ N+ + L + F+ G + CK+ + YGF+ +
Sbjct: 266 DPSTCRS----VYVGNIHTQVTELLLQEVFAGTGPVEGCKLFRK---EKSSYGFIHYFDR 424
Query: 171 DSAQNAIDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNE 230
SA AI LNG + + + V R++ + N++V +LS T+ L
Sbjct: 425 RSAALAILTLNGRHLFGQPIKVNWAYASGQREDT---SGHYNIFVGDLSPEVTDATLFAC 595
Query: 231 FGAYGTITSAVLMRD-ADGRSKCFGFVNFENAEDAAKAVEAL 271
F Y + + A +M D GRS+ FGFV+F + +DA A+ L
Sbjct: 596 FSVYPSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDL 721
Score = 55.8 bits (133), Expect = 2e-08
Identities = 45/176 (25%), Positives = 76/176 (42%)
Frame = +2
Query: 212 NVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEAL 271
+VYV N+ TE L+ F G + L R +GF+++ + AA A+ L
Sbjct: 284 SVYVGNIHTQVTELLLQEVFAGTGPVEGCKLFRK---EKSSYGFIHYFDRRSAALAILTL 454
Query: 272 NGKKVDDEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKL 331
NG+ + + V A +RE + G + N+++ +L +TD L
Sbjct: 455 NGRHLFGQPIKVNWAYASGQRE-DTSGHY---------------NIFVGDLSPEVTDATL 586
Query: 332 KEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEM 387
F + Y C+ D RV+ G SRG GFV+F + ++A A+ ++
Sbjct: 587 ------FACFSVYPSCS-----DARVMWDQKTGRSRGFGFVSFRSQQDAQSAINDL 721
Score = 44.7 bits (104), Expect = 5e-05
Identities = 23/61 (37%), Positives = 35/61 (56%)
Frame = +2
Query: 33 SLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDV 92
+++VGDL V D+ L+ F+ RV D T RS G+G+V+F + QDA A++
Sbjct: 539 NIFVGDLSPEVTDATLFACFSVYPSCSDARVMWDQKTGRSRGFGFVSFRSQQDAQSAIND 718
Query: 93 L 93
L
Sbjct: 719 L 721
>TC18542 similar to UP|Q93YF1 (Q93YF1) Nucleic acid binding protein, partial
(29%)
Length = 568
Score = 56.2 bits (134), Expect = 2e-08
Identities = 35/122 (28%), Positives = 61/122 (49%), Gaps = 4/122 (3%)
Frame = +2
Query: 71 RSLGYGYVNFTNPQDAARALDVLNFT--PMNNKSIRVMYSHRDPSSRKSGTANIFIKNLD 128
R+ GYG+V F + A R L N T P + R++++ +IF+ +L
Sbjct: 11 RAEGYGFVEFVSHAAAERVLHTYNGTLMPGTEQPFRLIWASSGGGGDAGPDHSIFVGDLA 190
Query: 129 KTIDHKALHDTFSS-FGQIMSCKIATDG-SGQSKGYGFVQFEAEDSAQNAIDKLNGMLIN 186
+ L +TF + + + K+ TD +G+SKGYGFV+F E A+ ++NG+ +
Sbjct: 191 PDVTDFLLQETFRTHYPSVRGAKVVTDPVTGRSKGYGFVKFSDEAQRNRAMTEMNGVYCS 370
Query: 187 DK 188
+
Sbjct: 371 SR 376
Score = 44.3 bits (103), Expect = 6e-05
Identities = 36/121 (29%), Positives = 60/121 (48%), Gaps = 6/121 (4%)
Frame = +2
Query: 316 NLYLKNLDDSITDEKLKEMF-SEFGTITSYKVCTMILFTDIRVIMRDP-NGVSRGSGFVA 373
++++ +L +TD L+E F + + ++ KV T DP G S+G GFV
Sbjct: 167 SIFVGDLAPDVTDFLLQETFRTHYPSVRGAKVVT------------DPVTGRSKGYGFVK 310
Query: 374 FSTPEEASRALGEMNGKMIVSKPLYVAVAQRKEDRRARLQAQFSQMRPV----AITPSVA 429
FS + +RA+ EMNG S+P+ ++ A K + Q Q++ + + A T VA
Sbjct: 311 FSDEAQRNRAMTEMNGVYCSSRPMRISAATPK--KTPSFQQQYAPTKAMYQYPAYTAPVA 484
Query: 430 P 430
P
Sbjct: 485 P 487
Score = 41.2 bits (95), Expect = 5e-04
Identities = 32/121 (26%), Positives = 61/121 (49%), Gaps = 3/121 (2%)
Frame = +2
Query: 156 SGQSKGYGFVQFEAEDSAQNAIDKLNGMLI-NDKQVFVGHFLRKQDRDNVLSKTKFNNVY 214
S +++GYGFV+F + +A+ + NG L+ +Q F + + ++++
Sbjct: 5 SARAEGYGFVEFVSHAAAERVLHTYNGTLMPGTEQPFRLIWASSGGGGDAGPD---HSIF 175
Query: 215 VKNLSESFTEDDLKNEFGA-YGTITSAVLMRD-ADGRSKCFGFVNFENAEDAAKAVEALN 272
V +L+ T+ L+ F Y ++ A ++ D GRSK +GFV F + +A+ +N
Sbjct: 176 VGDLAPDVTDFLLQETFRTHYPSVRGAKVVTDPVTGRSKGYGFVKFSDEAQRNRAMTEMN 355
Query: 273 G 273
G
Sbjct: 356 G 358
>AV780552
Length = 478
Score = 54.7 bits (130), Expect = 5e-08
Identities = 27/44 (61%), Positives = 36/44 (81%)
Frame = -2
Query: 598 DSAAKVTGMLLEMDQPEVLHLIESPDALKAKVAEAMEVLRNVSQ 641
D AA+VTG LLEMDQ +VLHL+ESP+ L AKVAEA + +++S+
Sbjct: 474 DHAAQVTGTLLEMDQTQVLHLLESPEPLPAKVAEAKDSPQDMSR 343
>TC10587 weakly similar to GB|AAC50895.1|1518802|HSU63289 CUG-BP/hNab50
{Homo sapiens;} , partial (19%)
Length = 482
Score = 54.3 bits (129), Expect = 6e-08
Identities = 26/76 (34%), Positives = 48/76 (62%), Gaps = 1/76 (1%)
Frame = +1
Query: 120 ANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATD-GSGQSKGYGFVQFEAEDSAQNAID 178
AN+FI ++ + + L + F FG+++S K+ D +G SK +GFV +++ ++AQ+AI
Sbjct: 13 ANLFIYHIPQEFGDQDLANAFQPFGRVISAKVFVDKATGVSKCFGFVSYDSPEAAQSAIS 192
Query: 179 KLNGMLINDKQVFVGH 194
+NG + K++ V H
Sbjct: 193 MMNGYQLGGKKLKVQH 240
Score = 52.4 bits (124), Expect = 2e-07
Identities = 24/63 (38%), Positives = 40/63 (63%), Gaps = 1/63 (1%)
Frame = +1
Query: 212 NVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRD-ADGRSKCFGFVNFENAEDAAKAVEA 270
N+++ ++ + F + DL N F +G + SA + D A G SKCFGFV++++ E A A+
Sbjct: 16 NLFIYHIPQEFGDQDLANAFQPFGRVISAKVFVDKATGVSKCFGFVSYDSPEAAQSAISM 195
Query: 271 LNG 273
+NG
Sbjct: 196 MNG 204
Score = 47.4 bits (111), Expect = 7e-06
Identities = 32/96 (33%), Positives = 45/96 (46%)
Frame = +1
Query: 314 GLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVA 373
G NL++ ++ D+ L F FG + S KV F D GVS+ GFV+
Sbjct: 10 GANLFIYHIPQEFGDQDLANAFQPFGRVISAKV-----FVD------KATGVSKCFGFVS 156
Query: 374 FSTPEEASRALGEMNGKMIVSKPLYVAVAQRKEDRR 409
+ +PE A A+ MNG + K L V Q K D +
Sbjct: 157 YDSPEAAQSAISMMNGYQLGGKKLKV---QHKRDNK 255
>TC11829 weakly similar to UP|NAM8_YEAST (Q00539) NAM8 protein, partial (4%)
Length = 738
Score = 53.1 bits (126), Expect = 1e-07
Identities = 58/225 (25%), Positives = 82/225 (35%), Gaps = 2/225 (0%)
Frame = +1
Query: 307 SVVDKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVS 366
+V + NLY+ L ++ D+ L ++F +FG I KV I G+S
Sbjct: 193 AVKKEIDDTNLYIGYLPPTLEDDGLIQLFQQFGEIVMAKV-----------IKDRMTGLS 339
Query: 367 RGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRKEDRRARLQAQFSQMRPVAITP 426
+G GFV ++ A+ A+ MNG + + + V VA +
Sbjct: 340 KGYGFVKYADITMANNAILAMNGYRLEGRTIAVRVAGK---------------------- 453
Query: 427 SVAPRMPLYPPGTPGLGQQFMYGQGPPAMMPPQAGFGYQQQLVPGMRPGGGPMPSYFVPM 486
P P+ PPG P G P P + QQQ G G P +Y
Sbjct: 454 ---PPQPVVPPGPPASGMPTY-----PVPSQPHGAYPSQQQYAAGGPLGVAPPTNYGGTP 609
Query: 487 VQQGQQGQRPGGRRGGPGQQPP--QQVPMMQQQMLPRQRVYRYPP 529
V P P PP P MQ Q +P V+ YPP
Sbjct: 610 VPWAPPVPPPYAPYAPP---PPGSTMYPPMQGQPMPPYGVH-YPP 732
Score = 47.8 bits (112), Expect = 6e-06
Identities = 26/73 (35%), Positives = 40/73 (54%), Gaps = 1/73 (1%)
Frame = +1
Query: 121 NIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDG-SGQSKGYGFVQFEAEDSAQNAIDK 179
N++I L T++ L F FG+I+ K+ D +G SKGYGFV++ A NAI
Sbjct: 220 NLYIGYLPPTLEDDGLIQLFQQFGEIVMAKVIKDRMTGLSKGYGFVKYADITMANNAILA 399
Query: 180 LNGMLINDKQVFV 192
+NG + + + V
Sbjct: 400 MNGYRLEGRTIAV 438
Score = 37.7 bits (86), Expect = 0.006
Identities = 21/67 (31%), Positives = 37/67 (54%), Gaps = 1/67 (1%)
Frame = +1
Query: 212 NVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDA-DGRSKCFGFVNFENAEDAAKAVEA 270
N+Y+ L + +D L F +G I A +++D G SK +GFV + + A A+ A
Sbjct: 220 NLYIGYLPPTLEDDGLIQLFQQFGEIVMAKVIKDRMTGLSKGYGFVKYADITMANNAILA 399
Query: 271 LNGKKVD 277
+NG +++
Sbjct: 400 MNGYRLE 420
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.134 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,941,109
Number of Sequences: 28460
Number of extensions: 120279
Number of successful extensions: 1086
Number of sequences better than 10.0: 148
Number of HSP's better than 10.0 without gapping: 847
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1021
length of query: 659
length of database: 4,897,600
effective HSP length: 96
effective length of query: 563
effective length of database: 2,165,440
effective search space: 1219142720
effective search space used: 1219142720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148486.10


