

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148485.6 + phase: 0
(380 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
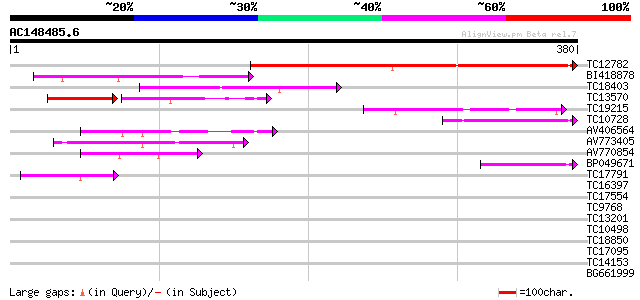
Score E
Sequences producing significant alignments: (bits) Value
TC12782 203 4e-53
BI418878 112 1e-25
TC18403 similar to UP|Q9LJE0 (Q9LJE0) Gb|AAC16072.1 (AT3g13510/M... 98 3e-21
TC13570 weakly similar to UP|Q9SL64 (Q9SL64) At2g20170 protein, ... 60 2e-14
TC19215 weakly similar to UP|Q9XIN9 (Q9XIN9) At2g27320 protein, ... 75 2e-14
TC10728 74 3e-14
AV406564 60 6e-10
AV773405 55 1e-08
AV770854 48 2e-06
BP049671 47 7e-06
TC17791 similar to UP|BAC99644 (BAC99644) Carboxyl-terminal prot... 42 2e-04
TC16397 UP|ACCD_LOTJA (Q9BBS1) Acetyl-coenzyme A carboxylase car... 29 1.5
TC17554 UP|CYB6_LOTJA (Q9BBQ6) Cytochrome b6, complete 29 1.5
TC9768 similar to GB|AAP68290.1|31711868|BT008851 At3g12156 {Ara... 28 1.9
TC13201 similar to UP|Q8LRL6 (Q8LRL6) Nam-like protein 9, partia... 28 2.5
TC10498 27 4.3
TC18850 similar to UP|Q41414 (Q41414) Epoxide hydrolase , parti... 27 4.3
TC17095 similar to UP|Q9FMC0 (Q9FMC0) Amino acid transporter-lik... 27 5.6
TC14153 weakly similar to UP|P93333 (P93333) PR10-1 protein (Cla... 27 5.6
BG661999 27 7.3
>TC12782
Length = 982
Score = 203 bits (516), Expect = 4e-53
Identities = 93/226 (41%), Positives = 149/226 (65%), Gaps = 7/226 (3%)
Frame = +2
Query: 162 KVDLPDDSTTAQIWLKAGN-GNEFESIEAGWMVNPGLYGNHDTRLFSYWTTDSYKSTGCF 220
K+ P++ + +QIW+ AG G + SIEAGW V+P LYG+++TRLF+YWT+D+Y++TGC+
Sbjct: 14 KIQQPNEFSLSQIWILAGAFGQDLNSIEAGWQVSPDLYGDNNTRLFTYWTSDAYQATGCY 193
Query: 221 DLTCSGFVQTSNTVALGGGINPISSDSGTQYELNI-----DEVGHWWLKENHDIPIGYWP 275
+L CSGF+Q +N +ALG I+P+S+ +QY+++I G+WW++ + +GYWP
Sbjct: 194 NLLCSGFIQINNDIALGASISPLSNYGSSQYDISILVWKDPNEGNWWMQFGNQHVLGYWP 373
Query: 276 VELFTSLKHSATLVQWGGQVFSSQVKKSPHTKTQMGSGHLADEKYGHACYMRNIRIKDNS 335
LF+ L SA++++WGG+V +S+ HT TQMGSGH +E +G A Y +NI+I D +
Sbjct: 374 APLFSYLTDSASMIEWGGEVVNSE-SDGQHTSTQMGSGHFPEEGFGKASYFKNIQIVDGN 550
Query: 336 LMLKYPESINVASQEPNCYSA-FNDEDVQEPTFYFGGPGQSSPSCP 380
+ P+ + ++ CY+ + FY+GGPG+ +P CP
Sbjct: 551 NKFRAPKDLGTYTEXNGCYNVKTGNAGDWGNYFYYGGPGR-NPXCP 685
>BI418878
Length = 609
Score = 112 bits (279), Expect = 1e-25
Identities = 64/179 (35%), Positives = 92/179 (50%), Gaps = 32/179 (17%)
Frame = +2
Query: 17 AGAISLSETEIEAKLKLH--NRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLKNHTI--- 71
A +S S ++E L+ N+P VKTI+S +GDIIDCV + KQPAFDHP LK+H I
Sbjct: 95 AARLSASRQKLEVNKHLNRLNKPPVKTIQSPDGDIIDCVHVSKQPAFDHPFLKDHKIQMR 274
Query: 72 --------------------------QTWHKSGRCPKGTVPIRRIQKQDLLRAATLDRFG 105
Q WH G+CPK T+P+RR +++D+LRA+++ R+G
Sbjct: 275 PSFHPEGLFDETKLSENKEKTSAPITQLWHSIGKCPKDTIPVRRTKEEDVLRASSVKRYG 454
Query: 106 LKQSSSFVNSKNTTISNFSKLSGSSNVVSED-HSGVHLATSGSNFIGAEADINVWNPKV 163
K+ S K S +++++ H G F GA+A INVW PK+
Sbjct: 455 RKKHRSIPKPK----------SAEPDLINQSGHQHAIAYVQGDKFYGAKATINVWEPKI 601
>TC18403 similar to UP|Q9LJE0 (Q9LJE0) Gb|AAC16072.1 (AT3g13510/MRP15_15),
partial (23%)
Length = 416
Score = 97.8 bits (242), Expect = 3e-21
Identities = 53/138 (38%), Positives = 82/138 (59%), Gaps = 3/138 (2%)
Frame = +2
Query: 88 RRIQKQDLLRAATLDRFGLKQSSSFVNSKNTT-ISNFSKLSGSSNVVSEDHSGVHLATSG 146
RR D+LRA +L FG K S+ T +S S + + + ++G+ + S
Sbjct: 2 RRSTVDDVLRAKSLYEFGKKPPGMLPLSRRTANVSPNDIFSNNDHEHAVAYTGLG-SDSP 178
Query: 147 SNFIGAEADINVWNPKVDLPDDSTTAQIWLKAG--NGNEFESIEAGWMVNPGLYGNHDTR 204
GA+A INVW P + + ++ + +QIWL +G +G++ SIEAGW V+P +YG+ R
Sbjct: 179 QEIYGAKASINVWEPSIQVMNEFSLSQIWLLSGFFDGSDLNSIEAGWQVSPQIYGDSRPR 358
Query: 205 LFSYWTTDSYKSTGCFDL 222
LF+YWT+DSY TGC++L
Sbjct: 359 LFTYWTSDSYGKTGCYNL 412
>TC13570 weakly similar to UP|Q9SL64 (Q9SL64) At2g20170 protein, partial
(13%)
Length = 505
Score = 60.1 bits (144), Expect(2) = 2e-14
Identities = 28/47 (59%), Positives = 35/47 (73%)
Frame = -3
Query: 26 EIEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLKNHTIQ 72
E E +LK N+P + TI + G I+DCVDI KQPAFDHP+LKNH +Q
Sbjct: 458 E*ERQLKHINKPPIVTINTT*GYIVDCVDIKKQPAFDHPLLKNHKLQ 318
Score = 35.4 bits (80), Expect(2) = 2e-14
Identities = 27/103 (26%), Positives = 45/103 (43%), Gaps = 3/103 (2%)
Frame = -1
Query: 76 KSGRCPKGTVPIRRIQKQDLLRAATLDRFGL---KQSSSFVNSKNTTISNFSKLSGSSNV 132
+ +CP GTVP+RR K+DL+R +L + + Q + + F ++ G+S
Sbjct: 238 RKDQCPVGTVPMRRTTKEDLIRGKSLFNYPI*TTGQPGTHIAEVTYGPFGFKRVYGASG- 62
Query: 133 VSEDHSGVHLATSGSNFIGAEADINVWNPKVDLPDDSTTAQIW 175
SGS +WNPKV D ++ + +W
Sbjct: 61 ------------SGS----------LWNPKVK-HDQTSNSHMW 2
>TC19215 weakly similar to UP|Q9XIN9 (Q9XIN9) At2g27320 protein, partial
(12%)
Length = 548
Score = 74.7 bits (182), Expect = 2e-14
Identities = 49/150 (32%), Positives = 73/150 (48%), Gaps = 14/150 (9%)
Frame = +1
Query: 238 GGINPISSDSGTQYELNIDE---VGHWWLKENHDI-PIGYWPVELFTSLKHSATLVQWGG 293
G + I SD + Y + + GHWWL +GYWP L T L H ++++++GG
Sbjct: 10 GPTSTIGSDYKSLYPFKVKQDQSTGHWWLIAGRGSWAVGYWPNPLLTHLSHGSSVIRYGG 189
Query: 294 QVFSSQVKKSPHTKTQMGSGHLADEKYGHACYMRNIRIKDNSLMLKYPE-SINVASQEPN 352
+ ++S SP MGSG L E Y ++ +M N+ I D++ Y E +N + N
Sbjct: 190 ETYASPHVMSP----PMGSGRLPRELYKNSAFMSNLEIIDSN----YNEVRVNPKYMKSN 345
Query: 353 CYSAFNDEDVQEP---------TFYFGGPG 373
C + N DVQ P F +GGPG
Sbjct: 346 CDTTPNCYDVQYPGYEGIVFAQAFLYGGPG 435
>TC10728
Length = 500
Score = 74.3 bits (181), Expect = 3e-14
Identities = 38/91 (41%), Positives = 55/91 (59%), Gaps = 1/91 (1%)
Frame = +1
Query: 291 WGGQVFSSQVKKSPHTKTQMGSGHLADEKYGHACYMRNIRIKDNSLMLKYPESINVASQE 350
WGG+V +++ HT TQMGSGH A++ YG A Y RNI I D + L+ ++I ++
Sbjct: 1 WGGEVVNTRANDE-HTSTQMGSGHFAEDGYGKARYFRNIEIVDVTNTLRSVQNITTIAET 177
Query: 351 PNCYSAFNDEDVQEPT-FYFGGPGQSSPSCP 380
NCY+ + T FY+GGPG+ +P CP
Sbjct: 178 SNCYNIQTSYSKEWGTYFYYGGPGK-NPQCP 267
>AV406564
Length = 416
Score = 60.1 bits (144), Expect = 6e-10
Identities = 47/152 (30%), Positives = 70/152 (45%), Gaps = 20/152 (13%)
Frame = +2
Query: 48 DIIDCVDIYKQPAFDHPVLKNHTIQTW-----------------HKSGRCPKGTVPI--R 88
D DCVD+YKQPAF HP+LKNH IQ + H RCP G VPI
Sbjct: 5 DDFDCVDLYKQPAFQHPLLKNHKIQLFPTFTMSTMQLGRDNKDKHSRKRCPMGKVPIFKT 184
Query: 89 RIQKQDLLRAATLDRF-GLKQSSSFVNSKNTTISNFSKLSGSSNVVSEDHSGVHLATSGS 147
I++Q + + + + GL Q S++ +F+ LS + N++
Sbjct: 185 NIREQKINMSTSKSQLDGLNQ-----YSQSRPRHHFATLSTTQNMM-------------- 307
Query: 148 NFIGAEADINVWNPKVDLPDDSTTAQIWLKAG 179
F GA A V+N + + + +QIW++ G
Sbjct: 308 -FHGASAQAAVYNLSLQ-ANQYSLSQIWIQGG 397
>AV773405
Length = 425
Score = 55.5 bits (132), Expect = 1e-08
Identities = 40/137 (29%), Positives = 66/137 (47%), Gaps = 6/137 (4%)
Frame = +2
Query: 30 KLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLKNHTIQTWHKSGRCPKGTVPI-- 87
+L+LH+ + + D DCVD+Y QPAF HP+LKNH IQ + R P+
Sbjct: 17 QLQLHD---YQNFQPSIDDSFDCVDMYNQPAFQHPLLKNHKIQLFPTCLRTHSKIDPVLG 187
Query: 88 RRIQKQDLLRAATLDRFGLKQSSSFVNSKNTTISNFSKLSGSSNVVSEDHSGVHLATSGS 147
+ ++ Q+ +R L + + ++++ K T S+ + N S+ G H AT +
Sbjct: 188 KAVKYQNFIRECPLGKVPILKTTT--RQKMVTKSSSKSQLDNFNENSQSKPGHHFATLET 361
Query: 148 N----FIGAEADINVWN 160
F G A I+ +N
Sbjct: 362 TKDMMFRGGSARISTYN 412
>AV770854
Length = 477
Score = 48.1 bits (113), Expect = 2e-06
Identities = 29/90 (32%), Positives = 48/90 (53%), Gaps = 8/90 (8%)
Frame = +1
Query: 48 DIIDCVDIYKQPAFDHPVLKNHTIQ---TWHKSGRCPKGTVPIRRIQKQDLLRA-----A 99
D DC+DIYKQPAF HP+LKNH IQ T +S + + + I+ ++ +R
Sbjct: 127 DDFDCIDIYKQPAFQHPLLKNHKIQLFPTCMRSRMQNRSSYLGKAIKDENSMRGCRSGKV 306
Query: 100 TLDRFGLKQSSSFVNSKNTTISNFSKLSGS 129
+ + ++Q +S + + NF++ S S
Sbjct: 307 PIFKTTMRQKLKTHSSSRSELDNFNQYSKS 396
>BP049671
Length = 488
Score = 46.6 bits (109), Expect = 7e-06
Identities = 24/66 (36%), Positives = 37/66 (55%), Gaps = 1/66 (1%)
Frame = -3
Query: 316 ADEKYGHACYMRNIRIKDNSLMLKYPESINVASQEPNCYSAFNDEDVQEPT-FYFGGPGQ 374
AD+ +G A Y RN+ I D L +I ++ NCY+ + + + T FY+GGPG
Sbjct: 486 ADDGFGKASYFRNLEIVDTDNSLSSVNNILTLAENTNCYNIKSSSNNKWGTYFYYGGPG- 310
Query: 375 SSPSCP 380
++P CP
Sbjct: 309 NNPQCP 292
>TC17791 similar to UP|BAC99644 (BAC99644) Carboxyl-terminal
proteinase-like, partial (4%)
Length = 544
Score = 42.0 bits (97), Expect = 2e-04
Identities = 23/71 (32%), Positives = 38/71 (53%), Gaps = 5/71 (7%)
Frame = -3
Query: 8 IMLLWAFFVAGAISLSETEIEAKLKLHNRPAVKTIKSEN-----GDIIDCVDIYKQPAFD 62
IM ++ F + ++S + ++ N+ ++ +N D D VD+Y QPAF
Sbjct: 527 IMAIFLFLLVSSVSSAMK*SGGRIHHQNKQQLQLHDYQNFQPSIDDAWDGVDMYNQPAFH 348
Query: 63 HPVLKNHTIQT 73
HP+LKNH IQ+
Sbjct: 347 HPLLKNHKIQS 315
>TC16397 UP|ACCD_LOTJA (Q9BBS1) Acetyl-coenzyme A carboxylase carboxyl
transferase subunit beta (ACCASE beta chain) , complete
Length = 1514
Score = 28.9 bits (63), Expect = 1.5
Identities = 17/55 (30%), Positives = 24/55 (42%)
Frame = +3
Query: 107 KQSSSFVNSKNTTISNFSKLSGSSNVVSEDHSGVHLATSGSNFIGAEADINVWNP 161
K ++ +N N+ K+ S + E H G HL S S+ I D WNP
Sbjct: 687 KYKHLWIECENCYGLNYKKILKSKMNICE-HCGYHLKMSSSDRIELSIDPGTWNP 848
>TC17554 UP|CYB6_LOTJA (Q9BBQ6) Cytochrome b6, complete
Length = 740
Score = 28.9 bits (63), Expect = 1.5
Identities = 12/41 (29%), Positives = 19/41 (46%), Gaps = 6/41 (14%)
Frame = +3
Query: 240 INPISSDSGTQYE------LNIDEVGHWWLKENHDIPIGYW 274
+NPI S +Y+ I + +WW + I +GYW
Sbjct: 240 VNPIGSSMVGEYDGLNDDPARISRLSYWWF*KTSRIDLGYW 362
>TC9768 similar to GB|AAP68290.1|31711868|BT008851 At3g12156 {Arabidopsis
thaliana;}, partial (51%)
Length = 865
Score = 28.5 bits (62), Expect = 1.9
Identities = 14/50 (28%), Positives = 23/50 (46%), Gaps = 2/50 (4%)
Frame = +2
Query: 185 ESIEAGWMVNPGLYGNHDTRLFSYWTTDSYKSTGCFDLTC--SGFVQTSN 232
+ I + W ++ G + NH T WT ++ + C C FV+ SN
Sbjct: 701 KKIASWWTISEGKHCNHGT*KSILWTKTTFSAARCETFVC**LAFVRKSN 850
>TC13201 similar to UP|Q8LRL6 (Q8LRL6) Nam-like protein 9, partial (22%)
Length = 465
Score = 28.1 bits (61), Expect = 2.5
Identities = 23/88 (26%), Positives = 40/88 (45%), Gaps = 6/88 (6%)
Frame = +3
Query: 46 NGDIIDCVDIYKQPAFDHPVL---KNHTIQTWHKSG---RCPKGTVPIRRIQKQDLLRAA 99
+GD I VD+YK +D P L K ++ + SG + KG+ R ++ +
Sbjct: 195 HGDPIGIVDVYKYEPWDLPCLSKMKTRDLEWYFYSGLDKKYGKGSSRTNRATEKGYWKTT 374
Query: 100 TLDRFGLKQSSSFVNSKNTTISNFSKLS 127
DR + ++ V K T + +F + S
Sbjct: 375 GKDR-PVNHANRTVGMKKTLVYHFGRAS 455
>TC10498
Length = 818
Score = 27.3 bits (59), Expect = 4.3
Identities = 15/35 (42%), Positives = 17/35 (47%)
Frame = +1
Query: 344 INVASQEPNCYSAFNDEDVQEPTFYFGGPGQSSPS 378
+N QEPN F Q PT FGGPG P+
Sbjct: 199 MNSFMQEPNPMQGFPFNGHQHPT--FGGPGMQLPA 297
>TC18850 similar to UP|Q41414 (Q41414) Epoxide hydrolase , partial (22%)
Length = 474
Score = 27.3 bits (59), Expect = 4.3
Identities = 11/29 (37%), Positives = 14/29 (47%)
Frame = -1
Query: 49 IIDCVDIYKQPAFDHPVLKNHTIQTWHKS 77
I+D + + HP NH IQ WH S
Sbjct: 186 IVDLMGPFLMEEVSHPFQHNHIIQKWHIS 100
>TC17095 similar to UP|Q9FMC0 (Q9FMC0) Amino acid transporter-like protein,
partial (17%)
Length = 751
Score = 26.9 bits (58), Expect = 5.6
Identities = 25/80 (31%), Positives = 31/80 (38%)
Frame = -3
Query: 107 KQSSSFVNSKNTTISNFSKLSGSSNVVSEDHSGVHLATSGSNFIGAEADINVWNPKVDLP 166
K S N N+ N SK S N SE G + T NF NP D
Sbjct: 347 KTSKQPNNPGNSKYPNSSKHCWS*NQNSEK*KGTNFLTQKKNFN---------NPFKD*I 195
Query: 167 DDSTTAQIWLKAGNGNEFES 186
++T W K+ NG+E S
Sbjct: 194 QLNSTRPSWEKSTNGSELRS 135
>TC14153 weakly similar to UP|P93333 (P93333) PR10-1 protein (Class 10 PR
protein), partial (97%)
Length = 754
Score = 26.9 bits (58), Expect = 5.6
Identities = 15/55 (27%), Positives = 27/55 (48%)
Frame = -1
Query: 134 SEDHSGVHLATSGSNFIGAEADINVWNPKVDLPDDSTTAQIWLKAGNGNEFESIE 188
SE G + S +NF +EA N+W+ + + +S W +G G E+++
Sbjct: 268 SELLDGSRASISFNNFNSSEAWNNLWDDGITINGESFVKPSWGYSGGGLILENVK 104
>BG661999
Length = 466
Score = 26.6 bits (57), Expect = 7.3
Identities = 15/58 (25%), Positives = 29/58 (49%), Gaps = 5/58 (8%)
Frame = +3
Query: 135 EDHSGVHLATSGSN-----FIGAEADINVWNPKVDLPDDSTTAQIWLKAGNGNEFESI 187
++H G L TS + ++G + P+V+ P+D ++A+ W G+ FE +
Sbjct: 267 KEHRGEVLQTSAKSKMMMIWVGKVMSV----PRVECPEDKSSARPWSHWSPGSPFERV 428
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.132 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,080,116
Number of Sequences: 28460
Number of extensions: 97684
Number of successful extensions: 469
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 458
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 459
length of query: 380
length of database: 4,897,600
effective HSP length: 92
effective length of query: 288
effective length of database: 2,279,280
effective search space: 656432640
effective search space used: 656432640
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC148485.6


