

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148484.8 + phase: 0
(398 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
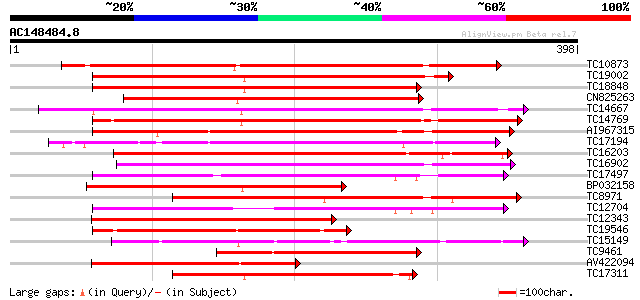
Score E
Sequences producing significant alignments: (bits) Value
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 273 3e-74
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 257 3e-69
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 246 6e-66
CN825263 246 6e-66
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 244 2e-65
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 235 1e-62
AI967315 229 6e-61
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 219 6e-58
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 208 1e-54
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 206 7e-54
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 195 1e-50
BP032158 181 2e-46
TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P... 179 9e-46
TC12704 UP|CAE45596 (CAE45596) S-receptor kinase-like protein 3,... 175 1e-44
TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine prote... 170 3e-43
TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine k... 169 6e-43
TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like pro... 165 1e-41
TC9461 similar to UP|O49840 (O49840) Protein kinase (AT2G02800/T... 162 9e-41
AV422094 157 3e-39
TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serin... 155 1e-38
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 273 bits (699), Expect = 3e-74
Identities = 150/315 (47%), Positives = 206/315 (64%), Gaps = 6/315 (1%)
Frame = +1
Query: 37 GMGPPGSMQSSPGLSLTLKGGTFTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEI 96
G G S SS G + +F + ELA AT+ F N+IG+GGFG V+KG L TG+ +
Sbjct: 358 GKGVSSSNGSSNGKTAA---ASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAV 528
Query: 97 AVKSLKAGSGQGEREFQAEIDIISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHL 156
AVK L QG +EF E+ ++S +HH +LV L+GYC G QR+LVYE++P +LE HL
Sbjct: 529 AVKQLSHDGRQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHL 708
Query: 157 ----HGKGVPTMDWPTRMRIALGSARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVA 212
H K ++W TRM++A+G+ARGL YLH P +I+RD+K+AN+L+D+ F K++
Sbjct: 709 FELSHDK--EPLNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLS 882
Query: 213 DFGLAKL-TTDTNTHVSTRVMGTFGYMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPL 271
DFGLAKL NTHVSTRVMGT+GY APEYA SGKLT KSD++SFGV+LLELLTG+R +
Sbjct: 883 DFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAI 1062
Query: 272 DLTNAM-DESLVDWARPLLSRALEEDGNFAELVDPFLEGNYDHQEMIRLAACAASSIRHS 330
D + +++LV WARP S + F +VDP L+G + + + + A A ++
Sbjct: 1063DTSRRPGEQNLVSWARPYFS----DRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQ 1230
Query: 331 AKKRSKMSQIVRALE 345
K R ++ IV ALE
Sbjct: 1231PKFRPLITDIVVALE 1275
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 257 bits (656), Expect = 3e-69
Identities = 131/256 (51%), Positives = 174/256 (67%), Gaps = 3/256 (1%)
Frame = +1
Query: 59 FTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDI 118
F+ +EL SAT F +N +G+GGFG V+ G L G +IAVK LK S + + EF E++I
Sbjct: 28 FSLKELHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKVWSNKADMEFAVEVEI 207
Query: 119 ISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPT--MDWPTRMRIALGS 176
++RV H++L+SL GYC G +R++VY+++PN +L HLHG+ +DW RM IA+GS
Sbjct: 208 LARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECLLDWNRRMNIAIGS 387
Query: 177 ARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFG 236
A G+ YLH +P IIHRDIKA+NVL+D F+A+VADFG AKL D THV+TRV GT G
Sbjct: 388 AEGIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPDGATHVTTRVKGTLG 567
Query: 237 YMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLD-LTNAMDESLVDWARPLLSRALEE 295
Y+APEYA GK E DVFSFG++LLEL +GK+PL+ L++ + S+ DWA PL
Sbjct: 568 YLAPEYAMLGKANECCDVFSFGILLLELASGKKPLEKLSSTVKRSINDWALPLACAK--- 738
Query: 296 DGNFAELVDPFLEGNY 311
F E DP L G Y
Sbjct: 739 --KFTEFADPRLNGEY 780
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 246 bits (627), Expect = 6e-66
Identities = 116/234 (49%), Positives = 171/234 (72%), Gaps = 3/234 (1%)
Frame = +2
Query: 59 FTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDI 118
FTY+EL +AT GF+++N +G+GGFG V+ G G +IAVK LKA + + E EF E+++
Sbjct: 266 FTYKELHAATGGFSDDNKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEV 445
Query: 119 ISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPT--MDWPTRMRIALGS 176
+ RV H++L+ L GYCV QR++VY+++PN +L HLHG+ ++W RM+IA+GS
Sbjct: 446 LGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQFAVEVQLNWQKRMKIAIGS 625
Query: 177 ARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFG 236
A G+ YLH + +P IIHRDIKA+NVL++ FE VADFG AKL + +H++TRV GT G
Sbjct: 626 AEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPLVADFGFAKLIPEGVSHMTTRVKGTLG 805
Query: 237 YMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLD-LTNAMDESLVDWARPLL 289
Y+APEYA GK++E DV+SFG++LLEL+TG++P++ L + ++ +WA PL+
Sbjct: 806 YLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGVKRTITEWAEPLI 967
>CN825263
Length = 663
Score = 246 bits (627), Expect = 6e-66
Identities = 121/214 (56%), Positives = 161/214 (74%), Gaps = 4/214 (1%)
Frame = +1
Query: 81 GFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRHLVSLVGYCVSGGQR 140
GFG V+KGIL G+++AVK LK +G REF AE++++SR+HHR+LV L+G C+ R
Sbjct: 1 GFGLVYKGILNDGRDVAVKILKRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQTR 180
Query: 141 MLVYEFVPNKTLEYHLHG--KGVPTMDWPTRMRIALGSARGLAYLHEDCSPRIIHRDIKA 198
L+YE VPN ++E HLHG K +DW RM+IALG+ARGLAYLHED +P +IHRD K+
Sbjct: 181 CLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRDFKS 360
Query: 199 ANVLIDDSFEAKVADFGLAKLTTDT-NTHVSTRVMGTFGYMAPEYASSGKLTEKSDVFSF 257
+N+L++ F KV+DFGLA+ D N H+ST VMGTFGY+APEYA +G L KSDV+S+
Sbjct: 361 SNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSDVYSY 540
Query: 258 GVMLLELLTGKRPLDLTNAM-DESLVDWARPLLS 290
GV+LLELLTG +P+DL+ E+LV WARP+L+
Sbjct: 541 GVVLLELLTGTKPVDLSQPPGQENLVTWARPILT 642
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 244 bits (622), Expect = 2e-65
Identities = 150/361 (41%), Positives = 214/361 (58%), Gaps = 17/361 (4%)
Frame = +2
Query: 21 PHPLMNSGEMSSNYSYGMG-PPGSMQSSPGLSLTLKGG------TFTYEELASATKGFAN 73
P+P + + S +YG G GS S+P T K + +EL T F +
Sbjct: 188 PYPSNENDHLKSPRNYGDGNSKGSKASAPVKHETQKAPPPIEVPALSLDELKEKTDNFGS 367
Query: 74 ENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSG-QGEREFQAEIDIISRVHHRHLVSLVG 132
+ +IG+G +G V+ L G +AVK L S + EF ++ ++SR+ + + V L G
Sbjct: 368 KALIGEGSYGRVYYATLNDGNAVAVKKLDVSSEPETNNEFLTQVSMVSRLKNDNFVELHG 547
Query: 133 YCVSGGQRMLVYEFVPNKTLEYHLHG-KGV------PTMDWPTRMRIALGSARGLAYLHE 185
YCV G R+L YEF +L LHG KGV PT+DW R+RIA+ +ARGL YLHE
Sbjct: 548 YCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLDWIQRVRIAVDAARGLEYLHE 727
Query: 186 DCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHV-STRVMGTFGYMAPEYAS 244
P IIHRDI+++NVLI + ++AK+ADF L+ D + STRV+GTFGY APEYA
Sbjct: 728 KVQPAIIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDMAARLHSTRVLGTFGYHAPEYAM 907
Query: 245 SGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAM-DESLVDWARPLLSRALEEDGNFAELV 303
+G+LT+KSDV+SFGV+LLELLTG++P+D T +SLV WA P LS + + V
Sbjct: 908 TGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQSLVTWATPRLS-----EDKVKQCV 1072
Query: 304 DPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKESMIKSPAP 363
DP L+G Y + + +LAA AA +++ A+ R MS +V+A L+ L ++ +PAP
Sbjct: 1073DPKLKGEYPPKGVAKLAAVAALCVQYEAEFRPNMSIVVKA------LQPLLKTPAAAPAP 1234
Query: 364 Q 364
+
Sbjct: 1235E 1237
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 235 bits (599), Expect = 1e-62
Identities = 133/306 (43%), Positives = 196/306 (63%), Gaps = 4/306 (1%)
Frame = +3
Query: 59 FTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDI 118
FT E + AT+ + + +IG+GGFG V++G L G+E+AVK + S QG REF E+++
Sbjct: 2133 FTLEYIEVATERY--KTLIGEGGFGSVYRGTLNDGQEVAVKVRSSTSTQGTREFDNELNL 2306
Query: 119 ISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGV--PTMDWPTRMRIALGS 176
+S + H +LV L+GYC Q++LVY F+ N +L+ L+G+ +DWPTR+ IALG+
Sbjct: 2307 LSAIQHENLVPLLGYCNESDQQILVYPFMSNGSLQDRLYGEPAKRKILDWPTRLSIALGA 2486
Query: 177 ARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLT-TDTNTHVSTRVMGTF 235
ARGLAYLH +IHRDIK++N+L+D S AKVADFG +K + +++VS V GT
Sbjct: 2487 ARGLAYLHTFPGRSVIHRDIKSSNILLDHSMCAKVADFGFSKYAPQEGDSYVSLEVRGTA 2666
Query: 236 GYMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAMDE-SLVDWARPLLSRALE 294
GY+ PEY + +L+EKSDVFSFGV+LLE+++G+ PL++ E SLV+WA P + R +
Sbjct: 2667 GYLDPEYYKTQQLSEKSDVFSFGVVLLEIVSGREPLNIKRPRTEWSLVEWATPYI-RGSK 2843
Query: 295 EDGNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLK 354
D E+VDP ++G Y + M R+ A + + R M IVR LE + +E+
Sbjct: 2844 VD----EIVDPGIKGGYHAEAMWRVVEVALQCLEPFSTYRPSMVAIVRELEDALIIENNA 3011
Query: 355 ESMIKS 360
+KS
Sbjct: 3012 SEYMKS 3029
>AI967315
Length = 1308
Score = 229 bits (584), Expect = 6e-61
Identities = 130/300 (43%), Positives = 190/300 (63%), Gaps = 4/300 (1%)
Frame = +1
Query: 59 FTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLK--AGSGQGEREFQAEI 116
F+YEEL AT GF++EN++G+GG+ V+KG L +G EIAVK L + E+EF EI
Sbjct: 112 FSYEELFHATNGFSSENMVGKGGYAEVYKGRLESGDEIAVKRLTRTCRDERKEKEFLTEI 291
Query: 117 DIISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGS 176
I V H +++ L+G C+ G LV+E ++ +H + + +DW TR +I LG+
Sbjct: 292 GTIGHVCHSNVMPLLGCCIDNGL-YLVFELSTVGSVASLIHDEKMAPLDWKTRYKIVLGT 468
Query: 177 ARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVS-TRVMGTF 235
ARGL YLH+ C RIIHRDIKA+N+L+ + FE +++DFGLAK TH S + GTF
Sbjct: 469 ARGLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPSQWTHHSIAPIEGTF 648
Query: 236 GYMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAMDESLVDWARPLLSRALEE 295
G++APEY G + EK+DVF+FGV LLE+++G++P+D +SL WA+P+LS+
Sbjct: 649 GHLAPEYYMHGVVDEKTDVFAFGVFLLEVISGRKPVD---GSHQSLHTWAKPILSK---- 807
Query: 296 DGNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRAL-EGDVSLEDLK 354
+LVDP LEG YD + R+A A+ IR S+ R MS+++ + EG++ E K
Sbjct: 808 -WEIEKLVDPRLEGCYDVTQFNRVAFAASLCIRASSTWRPTMSEVLEVMEEGEMDKERWK 984
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 219 bits (558), Expect = 6e-58
Identities = 134/325 (41%), Positives = 189/325 (57%), Gaps = 8/325 (2%)
Frame = +1
Query: 28 GEMSSNYSY---GMGPPGSMQSSPGLS--LTLKGGTFTYEELASATKGFANENIIGQGGF 82
G SS+ Y G PG+ S+ GL+ + K F+Y+ELA AT F+ +N IGQGGF
Sbjct: 946 GNASSSAEYETSGSSGPGTA-SATGLTSIMVAKSMEFSYQELAKATNNFSLDNKIGQGGF 1122
Query: 83 GYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRHLVSLVGYCVSGGQRML 142
G V+ L GK+ A+K + Q EF E+ +++ VHH +LV L+GYCV G L
Sbjct: 1123 GAVYYAEL-RGKKTAIKKMDV---QASTEFLCELKVLTHVHHLNLVRLIGYCVEGSL-FL 1287
Query: 143 VYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHEDCSPRIIHRDIKAANVL 202
VYE + N L +LHG G + W +R++IAL +ARGL Y+HE P IHRD+K+AN+L
Sbjct: 1288 VYEHIDNGNLGQYLHGSGKEPLPWSSRVQIALDAARGLEYIHEHTVPVYIHRDVKSANIL 1467
Query: 203 IDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMAPEYASSGKLTEKSDVFSFGVMLL 262
ID + KVADFGL KL N+ + TR++GTFGYM PEYA G ++ K DV++FGV+L
Sbjct: 1468 IDKNLRGKVADFGLTKLIEVGNSTLQTRLVGTFGYMPPEYAQYGDISPKIDVYAFGVVLF 1647
Query: 263 ELLTGKRPLDLTN---AMDESLVDWARPLLSRALEEDGNFAELVDPFLEGNYDHQEMIRL 319
EL++ K + T A + LV L+++ D +LVDP L NY ++++
Sbjct: 1648 ELISAKNAVLKTGELVAESKGLVALFEEALNKSDPCDA-LRKLVDPRLGENYPIDSVLKI 1824
Query: 320 AACAASSIRHSAKKRSKMSQIVRAL 344
A + R + R M +V AL
Sbjct: 1825 AQLGRACTRDNPLLRPSMRSLVVAL 1899
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 208 bits (529), Expect = 1e-54
Identities = 116/289 (40%), Positives = 177/289 (61%), Gaps = 9/289 (3%)
Frame = +2
Query: 74 ENIIGQGGFGYVHKGILPTGKEIAVKSLKA-GSGQGEREFQAEIDIISRVHHRHLVSLVG 132
ENIIG+GG G V++G +P G ++A+K L GSG+ + F+AEI+ + ++ HR+++ L+G
Sbjct: 2201 ENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRLLG 2380
Query: 133 YCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHEDCSPRII 192
Y + +L+YE++PN +L LHG + W R +IA+ +ARGL Y+H DCSP II
Sbjct: 2381 YVSNKDTNLLLYEYMPNGSLGEWLHGAKGGHLRWEMRYKIAVEAARGLCYMHHDCSPLII 2560
Query: 193 HRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVS-TRVMGTFGYMAPEYASSGKLTEK 251
HRD+K+ N+L+D FEA VADFGLAK D S + + G++GY+APEYA + K+ EK
Sbjct: 2561 HRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEK 2740
Query: 252 SDVFSFGVMLLELLTGKRPL-DLTNAMDESLVDWARPLLSRALEEDGNFAEL--VDPFLE 308
SDV+SFGV+LLEL+ G++P+ + + +D +V W +S + L VDP L
Sbjct: 2741 SDVYSFGVVLLELIIGRKPVGEFGDGVD--IVGWVNKTMSELSQPSDTALVLAVVDPRLS 2914
Query: 309 GNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRAL----EGDVSLEDL 353
G Y +I + A ++ R M ++V L + + S +DL
Sbjct: 2915 G-YPLTSVIHMFNIAMMCVKEMGPARPTMREVVHMLTNPPQSNTSTQDL 3058
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 206 bits (523), Expect = 7e-54
Identities = 112/282 (39%), Positives = 170/282 (59%), Gaps = 2/282 (0%)
Frame = +3
Query: 76 IIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRHLVSLVGYCV 135
++G GGFG V+ G + G ++A+K S QG EFQ EI+++S++ HRHLVSL+GYC
Sbjct: 12 LLGVGGFGKVYYGEVDGGTKVAIKRGNPLSEQGVHEFQTEIEMLSKLRHRHLVSLIGYCE 191
Query: 136 SGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHEDCSPRIIHRD 195
+ +LVY+ + TL HL+ P + W R+ I +G+ARGL YLH IIHRD
Sbjct: 192 ENTEMILVYDHMAYGTLREHLYKTQKPPLPWKQRLEICIGAARGLHYLHTGAKYTIIHRD 371
Query: 196 IKAANVLIDDSFEAKVADFGLAKL-TTDTNTHVSTRVMGTFGYMAPEYASSGKLTEKSDV 254
+K N+L+D+ + AKV+DFGL+K T NTHVST V G+FGY+ PEY +LT+KSDV
Sbjct: 372 VKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVKGSFGYLDPEYFRRQQLTDKSDV 551
Query: 255 FSFGVMLLELLTGKRPLDLTNAMDE-SLVDWARPLLSRALEEDGNFAELVDPFLEGNYDH 313
+SFGV+L E+L + L+ + A ++ SL +WA ++ G +++DP+L+G
Sbjct: 552 YSFGVVLFEILCARPALNPSLAKEQVSLAEWASHCYNK-----GILDQILDPYLKGKIAP 716
Query: 314 QEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKE 355
+ + A A + +R M ++ LE + L++ E
Sbjct: 717 ECFKKFAETAMKCVSDQGIERPSMGDVLWNLEFALQLQESAE 842
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 195 bits (496), Expect = 1e-50
Identities = 120/301 (39%), Positives = 166/301 (54%), Gaps = 9/301 (2%)
Frame = +1
Query: 59 FTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDI 118
F + ++SAT F+ N +G+GGFG V+KG+L G+EIAVK L SGQG EF+ EI +
Sbjct: 1636 FDFSTISSATNHFSLSNKLGEGGFGPVYKGLLANGQEIAVKRLSNTSGQGMEEFKNEIKL 1815
Query: 119 ISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSAR 178
I+R+ HR+LV L G V + NK ++ L +DW R++I G AR
Sbjct: 1816 IARLQHRNLVKLFGCSVHQDENSHA-----NKKMKILLDSTRSKLVDWNKRLQIIDGIAR 1980
Query: 179 GLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVST-RVMGTFGY 237
GL YLH+D RIIHRD+K +N+L+DD K++DFGLA++ T RVMGT+GY
Sbjct: 1981 GLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIFIGDQVEARTKRVMGTYGY 2160
Query: 238 MAPEYASSGKLTEKSDVFSFGVMLLELLTGKR------PLDLTNAMDESLVDW--ARPLL 289
M PEYA G + KSDVFSFGV++LE+++GK+ P N + + W RPL
Sbjct: 2161 MPPEYAVHGSFSIKSDVFSFGVIVLEIISGKKIGRFYDPHHHLNLLSHAWRLWIEERPL- 2337
Query: 290 SRALEEDGNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVS 349
ELVD L+ E++R A ++ + R M IV L G+
Sbjct: 2338 -----------ELVDELLDDPVIPTEILRYIHVALLCVQRRPENRPDMLSIVLMLNGEKE 2484
Query: 350 L 350
L
Sbjct: 2485 L 2487
>BP032158
Length = 555
Score = 181 bits (458), Expect = 2e-46
Identities = 92/184 (50%), Positives = 123/184 (66%), Gaps = 2/184 (1%)
Frame = +2
Query: 55 KGGTFTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQA 114
K G F+ ++ +AT F N IG+GGFG V+KG+L G IAVK L + S QG REF
Sbjct: 2 KTGYFSLRQIKAATNNFDPANKIGEGGFGPVYKGVLSEGDVIAVKQLSSKSKQGNREFIN 181
Query: 115 EIDIISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVP--TMDWPTRMRI 172
EI +IS + H +LV L G C+ G Q +LVYE++ N +L L G ++W TRM+I
Sbjct: 182 EIGMISALQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGNEEQKLNLNWRTRMKI 361
Query: 173 ALGSARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVM 232
+G A+GLAYLHE+ +I+HRDIKA NVL+D +AK++DFGLAKL + NTH+STR+
Sbjct: 362 CVGIAKGLAYLHEESRLKIVHRDIKATNVLLDKDLKAKISDFGLAKLDEEENTHISTRIA 541
Query: 233 GTFG 236
GT G
Sbjct: 542 GTIG 553
>TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P14.15
{Arabidopsis thaliana;}, partial (37%)
Length = 1079
Score = 179 bits (453), Expect = 9e-46
Identities = 99/251 (39%), Positives = 155/251 (61%), Gaps = 6/251 (2%)
Frame = +3
Query: 115 EIDIISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIAL 174
E+++++++ HR+LV L+G+ G +R+L+ E+VPN TL HL G +D+ R+ IA+
Sbjct: 6 EVELLAKIDHRNLVKLLGFIDKGNERILITEYVPNGTLREHLDGLRGKILDFNQRLEIAI 185
Query: 175 GSARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKL--TTDTNTHVSTRVM 232
A GL YLH +IIHRD+K++N+L+ +S AKVADFG A+L TH+ST+V
Sbjct: 186 DVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFARLGPVNGDQTHISTKVK 365
Query: 233 GTFGYMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAMDESL-VDWARPLLSR 291
GT GY+ PEY + +LT KSDV+SFG++LLE+LTG+RP++L A DE + + WA +
Sbjct: 366 GTVGYLDPEYMKTHQLTPKSDVYSFGILLLEILTGRRPVELKKAADERVTLRWAFRKYN- 542
Query: 292 ALEEDGNFAELVDPFLEG--NYD-HQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDV 348
+G+ EL+DP +E N D +M+ L+ A+ IR + + + A+ D
Sbjct: 543 ----EGSVVELMDPLMEEAVNADVLMKMLDLSFQCAAPIRTDRPNMKSVGEQLWAIRADY 710
Query: 349 SLEDLKESMIK 359
+E ++K
Sbjct: 711 LKSARRE*LVK 743
>TC12704 UP|CAE45596 (CAE45596) S-receptor kinase-like protein 3, complete
Length = 2481
Score = 175 bits (444), Expect = 1e-44
Identities = 108/313 (34%), Positives = 164/313 (51%), Gaps = 21/313 (6%)
Frame = +1
Query: 59 FTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDI 118
F + ++S T F+ N +G+GGFG V+KG+L G+EIAVK L SGQG EF+ E+ +
Sbjct: 1417 FDFSTISSTTNHFSESNKLGEGGFGPVYKGVLANGQEIAVKRLSNTSGQGMEEFKNEVKL 1596
Query: 119 ISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSAR 178
I+R+ HR+LV L+G + + +L+YEF+ N++L+Y +
Sbjct: 1597 IARLQHRNLVKLLGCSIHHDEMLLIYEFMHNRSLDYFIF--------------------- 1713
Query: 179 GLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVST-RVMGTFGY 237
D RIIHRD+K +N+L+D K++DFGLA++ T T RVMGT+GY
Sbjct: 1714 -------DSRLRIIHRDLKTSNILLDSEMNPKISDFGLARIFTGDQVEAKTKRVMGTYGY 1872
Query: 238 MAPEYASSGKLTEKSDVFSFGVMLLELLTGKR------PLDLTNAMDES------LVDWA 285
M+PEYA G + KSDVFSFGV++LE+++GK+ P N + S L+
Sbjct: 1873 MSPEYAVHGSFSVKSDVFSFGVIVLEIISGKKIGRFCDPHHHRNLLSHSSNFAVFLIKAL 2052
Query: 286 RPLLSRALEE--------DGNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKM 337
R + ++ + ELVD L+G E++R A ++ + R M
Sbjct: 2053 RICMFENVKNRKAWRLWIEERPLELVDELLDGLAIPTEILRYIHIALLCVQQRPEYRPDM 2232
Query: 338 SQIVRALEGDVSL 350
+V L G+ L
Sbjct: 2233 LSVVLMLNGEKEL 2271
>TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine protein kinase
Cdk9 (Cyclin-dependent kinase Cdk9) , partial (7%)
Length = 723
Score = 170 bits (431), Expect = 3e-43
Identities = 87/173 (50%), Positives = 116/173 (66%), Gaps = 1/173 (0%)
Frame = +3
Query: 58 TFTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEID 117
TF+YE L +ATK F N +G+GGFG V KG L G+EIAVK L S QG +F E
Sbjct: 204 TFSYETLVAATKNFHAVNKLGEGGFGPVFKGKLNDGREIAVKKLSRRSNQGRTQFINEAK 383
Query: 118 IISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHL-HGKGVPTMDWPTRMRIALGS 176
+++RV HR++VSL GYC G +++LVYE+VP ++L+ L + +DW R I G
Sbjct: 384 LLTRVQHRNVVSLFGYCAHGSEKLLVYEYVPRESLDKLLFRSQKKEQLDWKRRFDIISGV 563
Query: 177 ARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVST 229
ARGL YLHED IIHRDIKAAN+L+D+ + K+ADFGLA++ + THV+T
Sbjct: 564 ARGLLYLHEDSHDCIIHRDIKAANILLDEKWVPKIADFGLARIFPEDQTHVNT 722
>TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine kinase ,
partial (46%)
Length = 731
Score = 169 bits (429), Expect = 6e-43
Identities = 84/182 (46%), Positives = 125/182 (68%)
Frame = +1
Query: 59 FTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDI 118
++Y+++ AT+ F I+GQG FG V+K +PTG+ +AVK L S QGE EFQ E+ +
Sbjct: 199 YSYKDIQKATQNFTT--ILGQGSFGTVYKATMPTGEVVAVKVLAPNSKQGEHEFQTEVHL 372
Query: 119 ISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSAR 178
+ R+HHR+LV+LVG+CV GQR+LVY+F+ N +L L+G+ + W R++IA+ +
Sbjct: 373 LGRLHHRNLVNLVGFCVDKGQRILVYQFMSNGSLANLLYGE-EKELSWDERLQIAMDISH 549
Query: 179 GLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYM 238
G+ YLHE P +IHRD+K+AN+L+DDS A VADFGL+K + ++ + GT+GYM
Sbjct: 550 GIEYLHEGAVPPVIHRDLKSANILLDDSMRAMVADFGLSK--EEIFDGRNSGLKGTYGYM 723
Query: 239 AP 240
P
Sbjct: 724 DP 729
>TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like protein kinase
protein (AT3g17840/MEB5_6), partial (45%)
Length = 1489
Score = 165 bits (418), Expect = 1e-41
Identities = 108/297 (36%), Positives = 166/297 (55%), Gaps = 4/297 (1%)
Frame = +3
Query: 72 ANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRHLVSLV 131
A+ ++G+G FG +K +L TG +AVK LK + E+EF+ +I+ + + H+ LV L
Sbjct: 279 ASAEVLGKGTFGTAYKAVLETGLVVAVKRLKDVT-ISEKEFKDKIETVGAMDHQSLVPLR 455
Query: 132 GYCVSGGQRMLVYEFVPNKTLEYHLHGK---GVPTMDWPTRMRIALGSARGLAYLHEDCS 188
Y S +++LVY+++P +L LHG G ++W R IALG+ARG+ YLH
Sbjct: 456 AYYFSRDEKLLVYDYMPMGSLSALLHGNKGAGRTPLNWEIRSGIALGAARGIEYLHSQ-G 632
Query: 189 PRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMAPEYASSGKL 248
P + H +IKA+N+L+ S+EAKV+DFGLA L ++T RV G Y APE K+
Sbjct: 633 PNVSHGNIKASNILLTKSYEAKVSDFGLAHLVGPSST--PNRVAG---YRAPEVTDPRKV 797
Query: 249 TEKSDVFSFGVMLLELLTGKRPLD-LTNAMDESLVDWARPLLSRALEEDGNFAELVDPFL 307
++K+DV+SFGV+LLELLTGK P L N L W + L+ + EL L
Sbjct: 798 SQKADVYSFGVLLLELLTGKAPTHALLNEEGVDLPRWVQSLVREEWTSEVFDLEL----L 965
Query: 308 EGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKESMIKSPAPQ 364
+EM++L A +R ++++ R++E ++ +LKE + P Q
Sbjct: 966 RYQNVEEEMVQLLQLAVDCAAPYPDRRPSIAEVTRSIE-ELCRSNLKEDQDQIPHDQ 1133
>TC9461 similar to UP|O49840 (O49840) Protein kinase (AT2G02800/T20F6.6) ,
partial (37%)
Length = 484
Score = 162 bits (410), Expect = 9e-41
Identities = 81/146 (55%), Positives = 112/146 (76%), Gaps = 2/146 (1%)
Frame = +3
Query: 146 FVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHEDCSPRIIHRDIKAANVLIDD 205
F+P +LE HL +G + W RM++A+G+ARGL++LH + ++I+RD KA+N+L+D
Sbjct: 3 FMPKGSLENHLFRRGPQPLSWSVRMKVAIGAARGLSFLH-NAKSQVIYRDFKASNILLDA 179
Query: 206 SFEAKVADFGLAKL-TTDTNTHVSTRVMGTFGYMAPEYASSGKLTEKSDVFSFGVMLLEL 264
F AK++DFGLAK T THVST+VMGT GY APEY ++G+LT KSDV+SFGV+LLEL
Sbjct: 180 EFNAKLSDFGLAKAGPTGDRTHVSTQVMGTQGYAAPEYVATGRLTAKSDVYSFGVVLLEL 359
Query: 265 LTGKRPLDLTNA-MDESLVDWARPLL 289
L+G+R +D T A +D++LVDWARP L
Sbjct: 360 LSGRRAVDKTIAGVDQNLVDWARPYL 437
>AV422094
Length = 462
Score = 157 bits (397), Expect = 3e-39
Identities = 79/147 (53%), Positives = 105/147 (70%)
Frame = +2
Query: 58 TFTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEID 117
TF+Y EL +AT F +N +G+GGFG V+KGIL G IAVK L GS QG+ +F AEI
Sbjct: 11 TFSYSELKNATNDFNIDNKLGEGGFGPVYKGILNDGTVIAVKQLSLGSHQGKSQFIAEIA 190
Query: 118 IISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSA 177
IS V HR+LV L G C+ G +R+LVYE++ NK+L+ L GK + +++W TR I LG A
Sbjct: 191 TISAVQHRNLVKLYGCCIEGSKRLLVYEYLENKSLDQALFGKAL-SLNWSTRYDICLGVA 367
Query: 178 RGLAYLHEDCSPRIIHRDIKAANVLID 204
RGL YLHE+ RI+HRD+KA+N+L+D
Sbjct: 368 RGLTYLHEESRLRIVHRDVKASNILLD 448
>TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serine-threonine
kinase 3, partial (7%)
Length = 606
Score = 155 bits (392), Expect = 1e-38
Identities = 83/185 (44%), Positives = 117/185 (62%), Gaps = 13/185 (7%)
Frame = +3
Query: 115 EIDIISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPT----------M 164
E++I+S + H ++V L S +LVY+++ N +L+ LH K P +
Sbjct: 3 EVEILSNIRHNNIVKLQCCISSEESLLLVYDYLENLSLDRWLHKKSKPPTVSGSVQNNII 182
Query: 165 DWPTRMRIALGSARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTN 224
DWP R+ IA+G A+GL Y+H DCSP ++HRD+K +N+L+D F AKVADFGLA ++
Sbjct: 183 DWPRRLHIAIGVAQGLCYMHHDCSPPVVHRDLKPSNILLDSQFNAKVADFGLAMMSVKPE 362
Query: 225 THVS-TRVMGTFGYMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAMDE--SL 281
+ + V GTFGY+APEYA + ++ EK DV+SFGV+LLEL TGK N DE SL
Sbjct: 363 ELATMSAVAGTFGYIAPEYALTIRVNEKIDVYSFGVILLELTTGKE----ANHGDEYSSL 530
Query: 282 VDWAR 286
+WAR
Sbjct: 531 AEWAR 545
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.133 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,825,242
Number of Sequences: 28460
Number of extensions: 74123
Number of successful extensions: 813
Number of sequences better than 10.0: 303
Number of HSP's better than 10.0 without gapping: 640
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 647
length of query: 398
length of database: 4,897,600
effective HSP length: 92
effective length of query: 306
effective length of database: 2,279,280
effective search space: 697459680
effective search space used: 697459680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC148484.8


