

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148484.5 - phase: 0
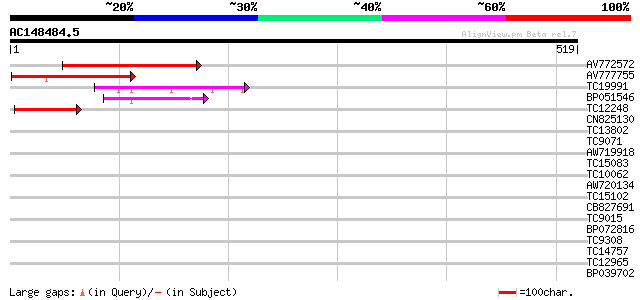
(519 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV772572 249 1e-66
AV777755 204 2e-53
TC19991 74 8e-14
BP051546 67 5e-12
TC12248 65 2e-11
CN825130 39 0.002
TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-... 39 0.002
TC9071 similar to UP|Q9LT85 (Q9LT85) Similarity to receptor prot... 37 0.006
AW719918 36 0.013
TC15083 similar to UP|GAR1_YEAST (P28007) snoRNP protein GAR1, p... 35 0.023
TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein... 35 0.023
AW720134 35 0.023
TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, ... 35 0.023
CB827691 35 0.039
TC9015 similar to UP|Q9SS90 (Q9SS90) F4P13.19 protein, partial (... 34 0.051
BP072816 34 0.051
TC9308 weakly similar to UP|Q9SQ55 (Q9SQ55) Nuclear RNA binding ... 33 0.086
TC14757 similar to UP|O04361 (O04361) Prohibitin, complete 33 0.15
TC12965 weakly similar to GB|AAP49518.1|31376383|BT008756 At3g62... 33 0.15
BP039702 33 0.15
>AV772572
Length = 491
Score = 249 bits (635), Expect = 1e-66
Identities = 110/127 (86%), Positives = 119/127 (93%)
Frame = -3
Query: 49 IALSPTTLLATNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKEIRKRVYVCPEP 108
IALSP TL+ATNRFVC ICN GFQRDQNLQLHRRGHNLPWKLRQRS+KE+RK+VY+CPE
Sbjct: 489 IALSPKTLMATNRFVCVICN*GFQRDQNLQLHRRGHNLPWKLRQRSNKEVRKKVYICPEK 310
Query: 109 TCVHHDPSRALGDLTGIKKHFCRKHGEKKWKCEKCSKKYAVQSDWKAHSKVCGSREYKCD 168
TCVHHD SRALGDLTGIKKHF RKHGEKKWKCEKCSKKYAV SDWKAH++ CG+REYKCD
Sbjct: 309 TCVHHDASRALGDLTGIKKHFSRKHGEKKWKCEKCSKKYAVPSDWKAHTQTCGTREYKCD 130
Query: 169 CGTVFSR 175
CGT+FSR
Sbjct: 129 CGTLFSR 109
>AV777755
Length = 373
Score = 204 bits (520), Expect = 2e-53
Identities = 98/117 (83%), Positives = 103/117 (87%), Gaps = 3/117 (2%)
Frame = +2
Query: 2 VDLDNVSTASGEASISSSGNNNIQSPIPKPT---KKKRNLPGMPDPEAEVIALSPTTLLA 58
VDLDNV TASGEAS+SSSGN + P T KKKRNLPGMPDP+AEVIALSP TL+A
Sbjct: 23 VDLDNVCTASGEASVSSSGNQIVPPPKAATTTTAKKKRNLPGMPDPDAEVIALSPKTLMA 202
Query: 59 TNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKEIRKRVYVCPEPTCVHHDP 115
TNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKE+RKRVYVCPEPTC HHDP
Sbjct: 203 TNRFVCEICNKGFQRDQNLQLHRRGHNLPWKLRQRSSKEVRKRVYVCPEPTCAHHDP 373
>TC19991
Length = 540
Score = 73.6 bits (179), Expect = 8e-14
Identities = 48/165 (29%), Positives = 75/165 (45%), Gaps = 23/165 (13%)
Frame = +1
Query: 78 QLHRRGHNLPWKLRQRSSKEI-------RKRVYVCPEPTC---VHHDPSRALGDLTGIKK 127
++H R H +K + +K + R+R + CP C H RAL + ++
Sbjct: 1 RMHMRAHGNQFKTAEALAKPLVEDAGGQRERRFSCPFKGCNRNKRHRRFRALKSVVCVRN 180
Query: 128 HFCRKHGEKKWKCEKCSKK--YAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFITHRA- 184
HF R H K + C++C K+ ++V SD K+H + C ++C CGT FSR+D H A
Sbjct: 181 HFKRSHCPKMYACDRCHKRKSFSVLSDLKSHMRHCSESRWRCSCGTTFSRKDKLFGHIAR 360
Query: 185 ---FCDALAEENAKSQNQAVGKANSESDSK-------VLTGDSLP 219
A+A + + + V + N E K L D LP
Sbjct: 361 FEGHVPAIALDEIGEKGKHVVEENEEDPIKEDELSNCFLNDDGLP 495
>BP051546
Length = 574
Score = 67.4 bits (163), Expect = 5e-12
Identities = 34/99 (34%), Positives = 48/99 (48%), Gaps = 3/99 (3%)
Frame = -2
Query: 87 PWKLRQRSSKEIRKRVYVCPEPTC---VHHDPSRALGDLTGIKKHFCRKHGEKKWKCEKC 143
P LR + + C P C + H ++ L D ++ H+ RKHG K + C KC
Sbjct: 564 PESLRGTQPTAMLRLPCYCCAPGCKNNIDHPRAKPLKDFRTLQTHYKRKHGIKPFMCRKC 385
Query: 144 SKKYAVQSDWKAHSKVCGSREYKCDCGTVFSRRDSFITH 182
K +AV+ DW+ H K CG Y C CG+ F + S H
Sbjct: 384 CKAFAVRGDWRTHEKNCGKLWY-CSCGSDFKHKRSLKDH 271
>TC12248
Length = 591
Score = 65.5 bits (158), Expect = 2e-11
Identities = 32/62 (51%), Positives = 43/62 (68%), Gaps = 1/62 (1%)
Frame = +3
Query: 5 DNVSTASGEASISSSGNNNIQSP-IPKPTKKKRNLPGMPDPEAEVIALSPTTLLATNRFV 63
DN + S+ ++ ++ +P P P KKKRN PG P+P+AEVIALSP TL+ATNRF+
Sbjct: 405 DNQMNQQQQHSLVAAAPSSSATPSAPPPQKKKRNQPGTPNPDAEVIALSPKTLMATNRFI 584
Query: 64 CE 65
CE
Sbjct: 585 CE 590
>CN825130
Length = 742
Score = 39.3 bits (90), Expect = 0.002
Identities = 39/122 (31%), Positives = 60/122 (48%), Gaps = 11/122 (9%)
Frame = +1
Query: 364 RQYVPTHQPPA---MSATALLQKAAQMG-AAATNASLLRGLGIVSSSASTSSGQQDSLHW 419
+Q +HQ A MSATALLQKAAQ+G ++T+ S L LG+ S ++ +
Sbjct: 370 QQQHQSHQTSASANMSATALLQKAAQIGTTSSTDPSFLGSLGLRSCNSPRQDHGNSNKFC 549
Query: 420 GLGPMEPEGSSLVSAGLGLGLPCDADSGLKELMLGTPSMFGPKQT-------TLDFLGLG 472
G+ SS+V+ G C+ ++ +L P+ Q+ T DFLG+G
Sbjct: 550 GM----YSSSSVVTTSHG----CETENYSDDLSQMPPAKRRHVQSEESAWGQTRDFLGVG 705
Query: 473 MA 474
+A
Sbjct: 706 VA 711
>TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-like
protein, partial (41%)
Length = 522
Score = 38.9 bits (89), Expect = 0.002
Identities = 39/156 (25%), Positives = 68/156 (43%), Gaps = 11/156 (7%)
Frame = +2
Query: 210 SKVLTGDSLPAVITTTAAAAATTPQSNSGVSSA--LETQKLDLPENPPQIIEEPQVVVTT 267
S +L L + +T + ++ P + +S+ T K P + P +
Sbjct: 38 STILPNKPLHSTLTKPPSQISSNPNKTTSTTSSPTSTTPKPSPSPAPSSTLRAPSSSPAS 217
Query: 268 TASALNASCSSSTSSKSNGCAATSTGVFASLFASSTA----SASASLQPQAP-AFSDLIR 322
+ A + + S SS S +ST + S S ++ S+S+S P P +FS L R
Sbjct: 218 ASPASSPTKSPKPSSPSASAPPSSTPLTPSTATSESSPTATSSSSSASPAPPTSFSALSR 397
Query: 323 SMG----CTDPRPTDFSAPPSSEAISLCLSTSHGSS 354
++ + P P S+PP S ++C+STS S+
Sbjct: 398 ALELKALASSPSPP--SSPPPSPPSAICMSTSLSSA 499
Score = 32.7 bits (73), Expect = 0.15
Identities = 39/148 (26%), Positives = 62/148 (41%), Gaps = 2/148 (1%)
Frame = +2
Query: 250 LPENPPQIIEEPQVVVTTTASALNASCSSSTSSKSNGCAATSTGVFASLFASSTASASAS 309
L + P QI P +TT+S +ST+ K + A S+ + A +SS ASAS +
Sbjct: 74 LTKPPSQISSNPNKTTSTTSSP------TSTTPKPSPSPAPSSTLRAP--SSSPASASPA 229
Query: 310 LQPQAPAFSDLIRSMGCTDPRPTD--FSAPPSSEAISLCLSTSHGSSIFGTGGQECRQYV 367
P P+P+ SAPPSS ++ +TS S +
Sbjct: 230 SSPTK-------------SPKPSSPSASAPPSSTPLTPSTATSESSPTATSSSSSASPAP 370
Query: 368 PTHQPPAMSATALLQKAAQMGAAATNAS 395
PT + + L +A ++ A A++ S
Sbjct: 371 PT-------SFSALSRALELKALASSPS 433
Score = 31.6 bits (70), Expect = 0.33
Identities = 21/69 (30%), Positives = 34/69 (48%), Gaps = 1/69 (1%)
Frame = +2
Query: 214 TGDSLPAVITTTAAAAATTPQSNSGVSSALETQKLDLPENPPQI-IEEPQVVVTTTASAL 272
T +S P +++++A+ P S S +S ALE + L +PP P + + S
Sbjct: 314 TSESSPTATSSSSSASPAPPTSFSALSRALELKALASSPSPPSSPPPSPPSAICMSTSLS 493
Query: 273 NASCSSSTS 281
+AS STS
Sbjct: 494 SASSVRSTS 520
>TC9071 similar to UP|Q9LT85 (Q9LT85) Similarity to receptor protein
kinase, partial (15%)
Length = 872
Score = 37.4 bits (85), Expect = 0.006
Identities = 46/176 (26%), Positives = 63/176 (35%)
Frame = +2
Query: 235 SNSGVSSALETQKLDLPENPPQIIEEPQVVVTTTASALNASCSSSTSSKSNGCAATSTGV 294
S S A E L PP P + + L AS S+S + + A G
Sbjct: 218 SKSSSVVAAEAAVARLLPPPPPAAHLPHPNHSAV*TKLAASSSNSPPTSTTPTATPKPGT 397
Query: 295 FASLFASSTASASASLQPQAPAFSDLIRSMGCTDPRPTDFSAPPSSEAISLCLSTSHGSS 354
A+STAS +A +Q +P P S PP+S A + LS S SS
Sbjct: 398 AKPTPANSTASDAAYIQTPT------------REPSPESTSTPPASPAKTATLSPSTASS 541
Query: 355 IFGTGGQECRQYVPTHQPPAMSATALLQKAAQMGAAATNASLLRGLGIVSSSASTS 410
+ PT P A + + +AT S + SSSA +S
Sbjct: 542 T-ASRNSHSST*TPTTSPALSRAKSSSSPTSTSSTSATTNSPASFRWMFSSSAPSS 706
>AW719918
Length = 506
Score = 36.2 bits (82), Expect = 0.013
Identities = 38/127 (29%), Positives = 58/127 (44%), Gaps = 8/127 (6%)
Frame = +1
Query: 223 TTTAAAAATTPQSNSGVSSALETQKLDLPENPPQIIEEPQVVVTTTASALNASCSSSTSS 282
T + + +T Q +S + + P N P ++ P + ++L AS S S S
Sbjct: 40 TPSLLSHSTPTQKHSDHNQNRHNHQWKPPLNSPHLLHPPPSTARSATTSLAAS-SRSASG 216
Query: 283 KSNGCAATST---GVFASLFASSTASASA-SLQPQAPAFSDLIRSM--GCTDPRPTDFSA 336
S+ ATST +S SST+SA+A SL P P + L+ + + P P+ SA
Sbjct: 217 TSSPFPATST*LSSTTSSASLSSTSSAAATSLTPATPPTATLVLRVLAPASSPSPSVVSA 396
Query: 337 --PPSSE 341
PS E
Sbjct: 397 SSTPSPE 417
>TC15083 similar to UP|GAR1_YEAST (P28007) snoRNP protein GAR1, partial (7%)
Length = 1367
Score = 35.4 bits (80), Expect = 0.023
Identities = 39/137 (28%), Positives = 54/137 (38%), Gaps = 9/137 (6%)
Frame = +2
Query: 251 PENPPQIIEEPQVVVTTTASALNASCSSSTSSKS-NGCAATSTGVFASLFASSTASASAS 309
P +PP+++ T+ ASC S N + TST A L S++A A S
Sbjct: 98 PLSPPKLLR-----AHPTSQ*SQASCG*ENHFPSLNQSSETSTPNMAQLSPSASAPAPPS 262
Query: 310 LQPQAPA-----FSDLIRSMGCTDPRPTDFSAPPSSEAISLCLSTSHGSSIFGTGGQECR 364
P AP+ F S P P S+PP+S +L L G T C
Sbjct: 263 SSPTAPSPTVPSFKTPPYSPTALSPSPPAKSSPPTSTTSALPLMAQPGVPSDATSPARCS 442
Query: 365 QY---VPTHQPPAMSAT 378
P+H+P S+T
Sbjct: 443 TPPV*SPSHKPGNGSST 493
>TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein 2, partial
(66%)
Length = 481
Score = 35.4 bits (80), Expect = 0.023
Identities = 30/97 (30%), Positives = 44/97 (44%)
Frame = -3
Query: 224 TTAAAAATTPQSNSGVSSALETQKLDLPENPPQIIEEPQVVVTTTASALNASCSSSTSSK 283
TT AA+ATT S + VS+A TTAS ++AS S S +S
Sbjct: 479 TTTAASATTSSSTATVSTA---------------------TTITTASTVSASSSISATSA 363
Query: 284 SNGCAATSTGVFASLFASSTASASASLQPQAPAFSDL 320
S+ T++ + AS +S+AS + S A + L
Sbjct: 362 SSP-TVTASAISASTITASSASTATSTASAASSLHSL 255
>AW720134
Length = 515
Score = 35.4 bits (80), Expect = 0.023
Identities = 43/145 (29%), Positives = 55/145 (37%), Gaps = 3/145 (2%)
Frame = +1
Query: 208 SDSKVLTGDSLPAVITTTAAAAATTPQSNSGVSSALETQKLDLPENPPQIIEEPQVVVTT 267
S S L +LP + T TA T P + +S E P PP P
Sbjct: 4 SSSSPLKHKTLPFIFTKTAQP--TPPPPTAPTTSTSEPSSTPSPPTPPNSTTPPLPEPPP 177
Query: 268 TASALNASCSSSTSSKSNGCAATSTGVFASLFASSTASASASLQPQAPAFSDLIRSMGCT 327
++SC+ AATST FA+ ASST + L +P RS G T
Sbjct: 178 PTPPTDSSCA----------AATSTPPFAT-SASSTPHTACKLSAPSPT----RRSYGTT 312
Query: 328 D---PRPTDFSAPPSSEAISLCLST 349
PT S+P S +L ST
Sbjct: 313 SALFATPTAPSSPRSIPDPALASST 387
>TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, partial
(68%)
Length = 1097
Score = 35.4 bits (80), Expect = 0.023
Identities = 44/161 (27%), Positives = 65/161 (40%), Gaps = 4/161 (2%)
Frame = +1
Query: 223 TTTAAAAATTPQSNSGVSSALETQKLDLPENPPQIIEEPQVVVTTTASALNASCSSSTSS 282
TT AA ++ P+S S + E L P +PP ++ +S S SSST+
Sbjct: 196 TTKAATNSSPPESRSTTPPSSEPTCLLAPASPP-------TPKSSHSSTPPPSRSSSTTP 354
Query: 283 KS-NGCAATSTGVFASLFASSTASASASLQPQAPAFSDLIRSMGCTDPRPTD---FSAPP 338
KS N + T+ ++TA A + P P S S + P T SAPP
Sbjct: 355 KSKNATSLTAPSCPPPASPAATAPAPSKTPPNPPTNSSKPSSCKSSLPNTTPSSLSSAPP 534
Query: 339 SSEAISLCLSTSHGSSIFGTGGQECRQYVPTHQPPAMSATA 379
S +TS S+ QE P PP ++++
Sbjct: 535 SP-------TTSPISTPSSPENQEKPASTPPSAPPRSTSSS 636
>CB827691
Length = 419
Score = 34.7 bits (78), Expect = 0.039
Identities = 17/23 (73%), Positives = 18/23 (77%)
Frame = +3
Query: 373 PAMSATALLQKAAQMGAAATNAS 395
P MSATALLQKAAQMGA T +
Sbjct: 219 PHMSATALLQKAAQMGATMTTTT 287
>TC9015 similar to UP|Q9SS90 (Q9SS90) F4P13.19 protein, partial (12%)
Length = 639
Score = 34.3 bits (77), Expect = 0.051
Identities = 20/65 (30%), Positives = 32/65 (48%)
Frame = +1
Query: 299 FASSTASASASLQPQAPAFSDLIRSMGCTDPRPTDFSAPPSSEAISLCLSTSHGSSIFGT 358
F +S S + S +P P+F+D +G P P S+ ++ +CL+ S + FG
Sbjct: 88 FGTSKPSHATSFEPSVPSFNDNSSVVGTAPPAP---SSAYDNQLCPICLTNSKDMA-FGC 255
Query: 359 GGQEC 363
G Q C
Sbjct: 256 GHQTC 270
>BP072816
Length = 492
Score = 34.3 bits (77), Expect = 0.051
Identities = 24/68 (35%), Positives = 35/68 (51%), Gaps = 1/68 (1%)
Frame = +1
Query: 278 SSTSSKSNGCAATSTGVFA-SLFASSTASASASLQPQAPAFSDLIRSMGCTDPRPTDFSA 336
SS+SS S+ + T + A S SS SA+A+ P P + S T +PT +A
Sbjct: 118 SSSSSSSSPLPSPPTTLHAPSTSPSSAPSATAAPPPSTPTPAANTSSKPSTSSKPTTSAA 297
Query: 337 PPSSEAIS 344
PP+S +S
Sbjct: 298 PPASSLLS 321
>TC9308 weakly similar to UP|Q9SQ55 (Q9SQ55) Nuclear RNA binding protein A,
partial (9%)
Length = 562
Score = 33.5 bits (75), Expect = 0.086
Identities = 24/61 (39%), Positives = 32/61 (52%), Gaps = 5/61 (8%)
Frame = -1
Query: 261 PQVVVTTTASALNASCSSSTSSKSNGCAAT-STGVFASLFAS----STASASASLQPQAP 315
P +T SA AS S+ T +S C +T STG ++ FAS ST S A+L P+
Sbjct: 433 PSTTSRSTGSASTASASTRTLCRSTLCRSTLSTGSASTAFASTRSVSTTSPPAALAPRTA 254
Query: 316 A 316
A
Sbjct: 253 A 251
Score = 30.0 bits (66), Expect = 0.96
Identities = 15/76 (19%), Positives = 41/76 (53%)
Frame = -1
Query: 222 ITTTAAAAATTPQSNSGVSSALETQKLDLPENPPQIIEEPQVVVTTTASALNASCSSSTS 281
++TT+ AA P++ + + + + LDL ++ + + + V + AS ++ CS+ ++
Sbjct: 298 VSTTSPPAALAPRTAAKATPWVPMKALDLIKSSALSVRDRECVPSAGASGTSSDCSNKST 119
Query: 282 SKSNGCAATSTGVFAS 297
++ A ++ + +S
Sbjct: 118 TRRITVTALNSSLLSS 71
>TC14757 similar to UP|O04361 (O04361) Prohibitin, complete
Length = 1141
Score = 32.7 bits (73), Expect = 0.15
Identities = 32/116 (27%), Positives = 49/116 (41%)
Frame = +1
Query: 230 ATTPQSNSGVSSALETQKLDLPENPPQIIEEPQVVVTTTASALNASCSSSTSSKSNGCAA 289
AT PQS S +S LP P P + + +S+ +++ SS+T S ++
Sbjct: 43 ATKPQSPSSQTSPAPPSASALPPPPSTPPSTPSMEASAPSSSTDSAASSTTPSAKEPISS 222
Query: 290 TSTGVFASLFASSTASASASLQPQAPAFSDLIRSMGCTDPRPTDFSAPPSSEAISL 345
S G ++S + + S AP S S P PT +A PS IS+
Sbjct: 223 -SHGSRNPTSSTSALAPTPSPPSPAPRISRW*TSPSVFSPAPTP-NASPSLSRISV 384
>TC12965 weakly similar to GB|AAP49518.1|31376383|BT008756 At3g62130
{Arabidopsis thaliana;}, partial (18%)
Length = 471
Score = 32.7 bits (73), Expect = 0.15
Identities = 41/129 (31%), Positives = 54/129 (41%), Gaps = 6/129 (4%)
Frame = +2
Query: 218 LPAVITTTAAAAATTPQSNSG------VSSALETQKLDLPENPPQIIEEPQVVVTTTASA 271
L TTT AA T+P+S S SS L + P P ++E TT ASA
Sbjct: 137 LTTTTTTTTAAPPTSPKSQSSHLYPPPPSSPLLRFNPNSPTTIPPLLES-----TTAASA 301
Query: 272 LNASCSSSTSSKSNGCAATSTGVFASLFASSTASASASLQPQAPAFSDLIRSMGCTDPRP 331
A+ S + G ++TS + S +ST S AS P AP+ T P
Sbjct: 302 --AAPPPSPPPNTTGSSSTSASLTTS---TSTTSNKASSAP-APSSK--------TSSTP 439
Query: 332 TDFSAPPSS 340
T + PSS
Sbjct: 440 TTSTRSPSS 466
>BP039702
Length = 553
Score = 32.7 bits (73), Expect = 0.15
Identities = 31/90 (34%), Positives = 48/90 (52%)
Frame = +3
Query: 236 NSGVSSALETQKLDLPENPPQIIEEPQVVVTTTASALNASCSSSTSSKSNGCAATSTGVF 295
N GVS++ T +PP+ ++E +TTTA+A A+C++ SS+ S +
Sbjct: 222 NHGVSASPSTAN-----SPPEKVKEE---ITTTATATAAACNAKLSSE----VPRSPTLP 365
Query: 296 ASLFASSTASASASLQPQAPAFSDLIRSMG 325
A + SS A A+ SL +APA + R MG
Sbjct: 366 AEIRLSSPAKATESL-TEAPAL--VARLMG 446
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.312 0.127 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,549,889
Number of Sequences: 28460
Number of extensions: 159926
Number of successful extensions: 1226
Number of sequences better than 10.0: 123
Number of HSP's better than 10.0 without gapping: 1138
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1204
length of query: 519
length of database: 4,897,600
effective HSP length: 94
effective length of query: 425
effective length of database: 2,222,360
effective search space: 944503000
effective search space used: 944503000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC148484.5


