

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148406.6 + phase: 0
(569 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
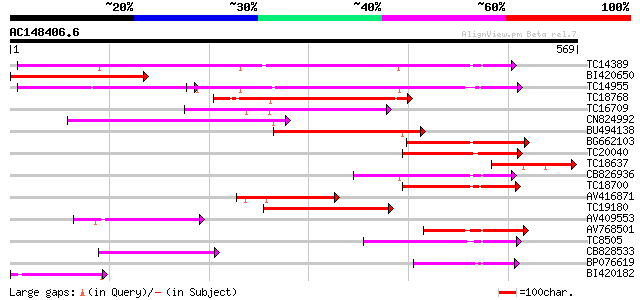
Score E
Sequences producing significant alignments: (bits) Value
TC14389 weakly similar to UP|Q9FNL8 (Q9FNL8) Peptide transporter... 328 1e-90
BI420650 236 1e-62
TC14955 UP|O22305 (O22305) Peptide transporter, complete 206 1e-53
TC18768 160 5e-40
TC16709 weakly similar to PIR|T47573|T47573 peptide transport-li... 153 6e-38
CN824992 131 3e-31
BU494138 125 1e-29
BG662103 125 1e-29
TC20040 similar to UP|Q9FM20 (Q9FM20) Nitrate transporter NTL1, ... 123 7e-29
TC18637 similar to UP|BAD12326 (BAD12326) NADH dehydrogenase sub... 109 1e-24
CB826936 103 1e-22
TC18700 similar to UP|Q9LYR6 (Q9LYR6) Peptide transporter-like p... 99 1e-21
AV416871 99 2e-21
TC19180 similar to PIR|T10255|T10255 nitrite transport protein, ... 96 1e-20
AV409553 94 5e-20
AV768501 83 1e-16
TC8505 similar to UP|Q8LG02 (Q8LG02) Nitrate transporter, partia... 82 3e-16
CB828533 77 8e-15
BP076619 74 8e-14
BI420182 71 5e-13
>TC14389 weakly similar to UP|Q9FNL8 (Q9FNL8) Peptide transporter, partial
(72%)
Length = 1990
Score = 328 bits (841), Expect = 1e-90
Identities = 183/517 (35%), Positives = 300/517 (57%), Gaps = 17/517 (3%)
Frame = +3
Query: 9 SLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVFGSLEVMAL 68
+LV++ +H +S+N ++N+ G + LVGA+I+D YL R+ T ++ + ++ +
Sbjct: 69 NLVVFLGSKLHEGTVASSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGM 248
Query: 69 VMITVQAALDNLHPKACGKS----SCVEGG--IAVMFYTSLCLYALGMGGVRGSLTAFGA 122
++T+ +L L P C + C + +FY +L + ALG GG + +++ GA
Sbjct: 249 CLLTLTVSLPALRPPPCAQGVENQDCPQASPWTRGIFYLALYIIALGAGGTKPNISTMGA 428
Query: 123 NQFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVASSIGF 182
+QFDE +P E +FFNW + S +G + T +V++ +W G+ + TV +
Sbjct: 429 DQFDEYEPKENTYKLSFFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLAFSI 608
Query: 183 VTLAIGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLPESNEQLYEV----YKDATIEK 238
+ +G PFYR K P SP+ R++QV V A K +P +QL+E+ Y + + +
Sbjct: 609 LVFLVGTPFYRHKLPSGSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNR 788
Query: 239 IVHTNQMRFLDKAAILQENSESQQPWKVCTVTQVEEVKILTRMLPILASTIVMNTCLAQL 298
I HT+ +RFLDKAA+ + PW++CTVTQVEE K +T+M+PIL +T++ T L Q
Sbjct: 789 IDHTSTLRFLDKAAV---KTGKTSPWRLCTVTQVEETKQMTKMVPILITTLIPCTMLIQA 959
Query: 299 QTFSVQQGNSMNLKLG-SFTVPASSIPVIPLIFLCTLIPIYELFFVPFIRKITHHPSGVT 357
T ++QG +++ +G +F +P + + I +I + T IPIY+ FVP IR+ T +P G+T
Sbjct: 960 HTLFIKQGTTLDRSMGPNFEIPPAGLSAILIISMLTSIPIYDRVFVPVIRRYTKNPRGIT 1139
Query: 358 QLQRVGVGLVLSAISMTIAGFIEVKRRDQGR------KDPSKPISLFWLSFQYAIFGIAD 411
LQR+G GL++ + M IA E KR R + P+S+F L QYA+ G+A+
Sbjct: 1140MLQRLGAGLLMYVLIMVIAWLTERKRLRVAREKHLLGQHDIIPLSIFILIPQYALTGVAE 1319
Query: 412 MFTLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSKQGWL 471
F + ++FFY +AP MKSL S++ S++LG FLS+ ++ + +TK+ S QGW+
Sbjct: 1320NFAEIAKMDFFYYQAPEGMKSLGISYSTTSVALGCFLSSFLLSTVADITKK--HSHQGWI 1493
Query: 472 QGFDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKY 508
+LN + L+ +Y F+ ILS LNF FL A ++ Y
Sbjct: 1494LD-NLNISRLDYYYAFMVILSFLNFLCFLVTAKFFDY 1601
>BI420650
Length = 576
Score = 236 bits (601), Expect = 1e-62
Identities = 117/139 (84%), Positives = 129/139 (92%)
Frame = +1
Query: 1 MGFVTNMVSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVF 60
MGFV NMVSLVLYFI VMHFDL+SSANTLTNFMGSTFLLSLVG FISDTYLNR TTCL+F
Sbjct: 160 MGFVANMVSLVLYFIMVMHFDLASSANTLTNFMGSTFLLSLVGGFISDTYLNRLTTCLLF 339
Query: 61 GSLEVMALVMITVQAALDNLHPKACGKSSCVEGGIAVMFYTSLCLYALGMGGVRGSLTAF 120
GSLEV+ALVM+TVQA LD+LHP+ACGKSSC++GGIAVM YTSL L ALG+GGVRG++ AF
Sbjct: 340 GSLEVLALVMLTVQAGLDSLHPEACGKSSCIKGGIAVMLYTSLGLLALGLGGVRGAMVAF 519
Query: 121 GANQFDEKDPNEAKALATF 139
GA+QFDEKDP EAKALATF
Sbjct: 520 GADQFDEKDPVEAKALATF 576
>TC14955 UP|O22305 (O22305) Peptide transporter, complete
Length = 2300
Score = 206 bits (523), Expect = 1e-53
Identities = 127/353 (35%), Positives = 200/353 (55%), Gaps = 16/353 (4%)
Frame = +1
Query: 178 SSIGFVTLAI----GKPFYRIKT-PGESPILRIIQVIVVAFKNRKLQLPESNEQLYEV-- 230
S+IGF+ +I G P YR K+ SP + I+V +VAF+NRKLQLP + +L+E
Sbjct: 682 SAIGFLLSSIIFFWGSPVYRHKSRQARSPSMNFIRVPLVAFRNRKLQLPCNPSELHEFQL 861
Query: 231 --YKDATIEKIVHTNQMRFLDKAAILQENSESQQPWKVCTVTQVEEVKILTRMLPILAST 288
Y + KI HT+ FLD+AAI + N++ P CTVTQVE K++ M I
Sbjct: 862 NYYISSGARKIHHTSHFSFLDRAAIRESNTDLSNP--PCTVTQVEGTKLVLGMFQIWLLM 1035
Query: 289 IVMNTCLAQLQTFSVQQGNSMNLKLG-SFTVPASSIPVIPLIFLCTLIPIYELFFVPFIR 347
++ C A T V+QG +M+ LG F +PA+S+ ++ +PIY+ +F+PF+R
Sbjct: 1036 LIPTNCWALESTIFVRQGTTMDRTLGPKFRLPAASLWCFIVLTTLICLPIYDHYFIPFMR 1215
Query: 348 KITHHPSGVTQLQRVGVGLVLSAISMTIAGFIEVKRRDQGRK------DPSKPISLFWLS 401
+ T + G+ LQRVG+G+ + I+M + +E +R +K + + P+S+FWL
Sbjct: 1216 RRTGNHRGIKLLQRVGIGMAIQVIAMAVTYAVETQRMSVIKKHHIVGPEETVPMSIFWLL 1395
Query: 402 FQYAIFGIADMFTLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTK 461
Q I G+++ F G+LEFFY ++P MK L T+ ++ G +++T V +I+
Sbjct: 1396 PQNIILGVSNAFLATGMLEFFYDQSPEEMKGLGTTLCTSCVAAGSYINTFLVTMID---- 1563
Query: 462 RITPSKQGWLQGFDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYKSEEDS 514
K W+ G +LN ++L+ +Y FL ++S LNF FL+ +S Y YK E S
Sbjct: 1564 -----KLNWI-GNNLNDSHLDYYYAFLFVISALNFGVFLWVSSGYIYKKENTS 1704
Score = 90.1 bits (222), Expect = 9e-19
Identities = 55/184 (29%), Positives = 91/184 (48%), Gaps = 2/184 (1%)
Frame = +3
Query: 9 SLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVFGSLEVMAL 68
+LV + ++ D+ SS + N+ G L ++GA+I+DTY RF T + + L
Sbjct: 183 NLVNFMTTQLNKDVVSSITSFNNWSGLATLTPILGAYIADTYTGRFWTITFSLLIYAIGL 362
Query: 69 VMITVQAALDNLHPKACGKSSCVEGGI--AVMFYTSLCLYALGMGGVRGSLTAFGANQFD 126
V++ + L +L P AC C E +FYTSL A+G G V+ +++ FGA+QFD
Sbjct: 363 VLLVLTTTLKSLRP-ACENGICREASNLQVALFYTSLYTIAVGSGAVKPNMSTFGADQFD 539
Query: 127 EKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVASSIGFVTLA 186
+ E + +FFNW + GS++ +V++ + +G I S L
Sbjct: 540 DFRHEEKEQKVSFFNWWAFNGACGSLMATLFVVYIQEKNGXGFGLQFICYWFSTFLYYLF 719
Query: 187 IGKP 190
+G P
Sbjct: 720 LGFP 731
>TC18768
Length = 738
Score = 160 bits (405), Expect = 5e-40
Identities = 91/204 (44%), Positives = 135/204 (65%), Gaps = 4/204 (1%)
Frame = +2
Query: 205 IIQVIVVAFKNRKLQLPESNEQLYEVYKDATIEKIVHTNQMRFLDKAAILQENSES---Q 261
+++ + + F RK P SN Q+ ++ D VHT++ RFLDKA I E + S +
Sbjct: 137 LLKFLWLHFFKRKHIYP-SNPQM--LHGDQNNVGQVHTDKFRFLDKACIRVEEAGSNTKK 307
Query: 262 QPWKVCTVTQVEEVKILTRMLPILASTIVMNTCLAQLQTFSVQQGNSMNLKLG-SFTVPA 320
W++C+V QVE+VKIL ++PI + TIV NT LAQLQTFSVQQG++M+ L SF +P
Sbjct: 308 SSWRLCSVGQVEQVKILLSVIPIFSCTIVFNTILAQLQTFSVQQGSAMDTHLTKSFHIPP 487
Query: 321 SSIPVIPLIFLCTLIPIYELFFVPFIRKITHHPSGVTQLQRVGVGLVLSAISMTIAGFIE 380
+S+ IP I L ++P+Y+ FFVPF R+IT H SG++ L+R+G GL L+ SM A +E
Sbjct: 488 ASLQSIPYILLIIVVPLYDTFFVPFARRITGHESGISPLRRIGFGLFLATFSMVSAALLE 667
Query: 381 VKRRDQGRKDPSKPISLFWLSFQY 404
KRRD+ + +K +S+FW++ Q+
Sbjct: 668 KKRRDEA-LNHNKTLSIFWITPQF 736
>TC16709 weakly similar to PIR|T47573|T47573 peptide transport-like protein
- Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(41%)
Length = 712
Score = 153 bits (387), Expect = 6e-38
Identities = 79/217 (36%), Positives = 129/217 (59%), Gaps = 9/217 (4%)
Frame = +1
Query: 176 VASSIGFVTLAIGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQLPESNEQLYEVYKDAT 235
VA ++ V+ G YR + PG SP+ R+ QV+V + + +Q P+ LYE+ +
Sbjct: 7 VAMAVAVVSFFSGTRLYRNQKPGGSPLTRMCQVVVASMRKCGVQAPDDKSLLYEIADTES 186
Query: 236 I----EKIVHTNQMRFLDKAAILQENSE---SQQPWKVCTVTQVEEVKILTRMLPILAST 288
K+ HTN++ F DKAA++ ++S+ S PW++CT TQVEE+K + RMLP+ A+
Sbjct: 187 AIKGSRKLDHTNELSFFDKAAVVVQSSDQKDSVDPWRLCTETQVEELKSILRMLPVWATG 366
Query: 289 IVMNTCLAQLQTFSVQQGNSMNLKLG--SFTVPASSIPVIPLIFLCTLIPIYELFFVPFI 346
I+ T Q+ T V QG +MN +G +F +P +++ + + + +P+Y+ VP
Sbjct: 367 IIFATVYGQMSTLFVLQGQTMNTHVGNSNFKIPPAALSIFDTLSVIFWVPVYDWIIVPIA 546
Query: 347 RKITHHPSGVTQLQRVGVGLVLSAISMTIAGFIEVKR 383
RK + H +G+TQLQR+G+GL +S +M A +EV R
Sbjct: 547 RKFSGHKNGLTQLQRMGIGLFISIFAMVAAAILEVIR 657
>CN824992
Length = 708
Score = 131 bits (330), Expect = 3e-31
Identities = 80/234 (34%), Positives = 123/234 (52%), Gaps = 11/234 (4%)
Frame = +2
Query: 59 VFGSLEVMALVMITVQAALDNLHPKACGKSS--CVEGGI--AVMFYTSLCLYALGMGGVR 114
+F + V+ L +++ L L P CG C +FY S+ L ALG GG +
Sbjct: 5 IFQVIFVVGLASLSLTTYLFLLEPNGCGDKELPCTSHSSYQTALFYVSIYLIALGNGGYQ 184
Query: 115 GSLTAFGANQFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFII 174
++ FGA+QFDE+DP E + +FF++ L+ LGS+ T + + W GF+
Sbjct: 185 PNIATFGADQFDEEDPKEQHSKISFFSYFYLALNLGSLFSNTILNYFEDDGLWTLGFWAS 364
Query: 175 TVASSIGFVTLAIGKPFYRIKTPGESPILRIIQVIVVAFKNRKLQL-PESNEQLYEVYKD 233
++ + V G P+YR PG +P+ R QV V A + K+++ P +QL+EV +
Sbjct: 365 AGSAFLALVLFLCGTPWYRYFKPGGNPVPRFCQVFVAAMRKWKVKVSPGEEDQLHEVEEC 544
Query: 234 ATI--EKIVHTNQMRFLDKAAILQENSESQQP----WKVCTVTQVEEVKILTRM 281
+T K+ HT RFLDKAA++ Q+ W + TVTQVEEVK + R+
Sbjct: 545 STNGGRKMFHTEGFRFLDKAALITSQESKQENECSLWHLSTVTQVEEVKCILRL 706
>BU494138
Length = 518
Score = 125 bits (315), Expect = 1e-29
Identities = 64/158 (40%), Positives = 98/158 (61%), Gaps = 5/158 (3%)
Frame = +2
Query: 265 KVCTVTQVEEVKILTRMLPILASTIVMNTCLAQLQTFSVQQGNSMNLKLGS-FTVPASSI 323
+V ++ QVEE+K L R+ PI A+ I+ T +AQ TF+V Q M+ +GS F +PA S+
Sbjct: 11 RVVSIQQVEEIKCLARIFPIWAAGILGFTAMAQQGTFTVSQAMKMDRHIGSKFQIPAGSL 190
Query: 324 PVIPLIFLCTLIPIYELFFVPFIRKITHHPSGVTQLQRVGVGLVLSAISMTIAGFIEVKR 383
VI I + +P Y+ FFVP +R+IT H G+T LQR+G+G+V S +SM +AG +E R
Sbjct: 191 GVISFITIGLWVPFYDRFFVPAVRRITKHEGGITLLQRIGIGMVFSVLSMIVAGLVEKVR 370
Query: 384 RDQGRKDPS----KPISLFWLSFQYAIFGIADMFTLVG 417
R +P+ P+S+ WL+ Q + G+ + F +G
Sbjct: 371 RGVANSNPNPLGIAPMSVMWLAPQLVLMGLCEAFNAIG 484
>BG662103
Length = 387
Score = 125 bits (315), Expect = 1e-29
Identities = 57/123 (46%), Positives = 85/123 (68%)
Frame = +1
Query: 399 WLSFQYAIFGIADMFTLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINT 458
W++FQY G AD+FTL GLLEFF+ EAP M+SL+TS ++ S+++GY+LS+ V+++N+
Sbjct: 19 WIAFQYLFLGSADLFTLAGLLEFFFSEAPIRMRSLATSLSWASLAIGYYLSSAIVSIVNS 198
Query: 459 VTKRITPSKQGWLQGFDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYKSEEDSNNSK 518
VT + S + WL G +LN +L FYW + +LS LNF ++LYWA+ YKY+ +N
Sbjct: 199 VTGK--GSHKPWLSGANLNHYHLERFYWLMCLLSGLNFLHYLYWAARYKYRRRGTANE*S 372
Query: 519 ELG 521
G
Sbjct: 373 NCG 381
>TC20040 similar to UP|Q9FM20 (Q9FM20) Nitrate transporter NTL1, partial
(20%)
Length = 578
Score = 123 bits (309), Expect = 7e-29
Identities = 58/122 (47%), Positives = 85/122 (69%), Gaps = 2/122 (1%)
Frame = +3
Query: 395 ISLFWLSFQYAIFGIADMFTLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVN 454
++ W++FQY G AD+FTL G++EFF+ EAP +M+SL+T+ ++ S+S+GYFLSTV V+
Sbjct: 3 MTFLWVAFQYLFLGSADLFTLAGMMEFFFTEAPWSMRSLATALSWASLSMGYFLSTVLVS 182
Query: 455 VINTVTKRI--TPSKQGWLQGFDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYKSEE 512
+IN VT TP WL G +LN +L FYW + +LS LNF ++L+WA+ YKY+
Sbjct: 183 IINKVTGAFGHTP----WLLGSNLNHYHLERFYWLMCVLSGLNFIHYLFWANSYKYRGSP 350
Query: 513 DS 514
S
Sbjct: 351 RS 356
>TC18637 similar to UP|BAD12326 (BAD12326) NADH dehydrogenase subunit 2,
partial (5%)
Length = 506
Score = 109 bits (273), Expect = 1e-24
Identities = 60/101 (59%), Positives = 70/101 (68%), Gaps = 16/101 (15%)
Frame = +1
Query: 484 FYWFLAILSCLNFFNFLYWASWYKYKSEEDS---NNSKELGETHLLMVGGRKHDDK---- 536
FYWFLAILSCLNFF+FLYWAS Y+Y+SE+ S N K + ET +MV K DK
Sbjct: 1 FYWFLAILSCLNFFDFLYWASCYQYQSEDHSCSKLNVKAVAETTAIMVVDEKKHDKEDMK 180
Query: 537 ---------AKAKESSQTSEANTEGPSSSDETDDGKEKERH 568
AKAKES+ TSEA+TEGPSSSDETDDG+ KER+
Sbjct: 181 DHGSTPDLRAKAKESNPTSEAHTEGPSSSDETDDGQPKERN 303
>CB826936
Length = 596
Score = 103 bits (256), Expect = 1e-22
Identities = 61/169 (36%), Positives = 98/169 (57%), Gaps = 6/169 (3%)
Frame = +3
Query: 346 IRKITHHPSGVTQLQRVGVGLVLSAISMTIAGFIEVKRRDQGRK------DPSKPISLFW 399
+R+ T +P G+ LQR+G GLV+ I+M A E KR R+ P+++F
Sbjct: 6 MRQYTKNPRGIAMLQRLGAGLVMYIITMVTAWLSERKRLSVAREYNLLGQHDKIPLTIFI 185
Query: 400 LSFQYAIFGIADMFTLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTV 459
L Q+A+ G+A+ F + +L+FFY +AP MKSL S++ S +LG F S+ ++ + +
Sbjct: 186 LLPQFALTGVANNFVDIAMLDFFYDQAPEGMKSLGISYSTTSTALGCFFSSFLLSTVADL 365
Query: 460 TKRITPSKQGWLQGFDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKY 508
TK+ +GW+ +LN ++L+ +Y F+AILS NF FL A + Y
Sbjct: 366 TKK--HGHKGWVLD-NLNVSHLDYYYAFMAILSVANFLCFLVAAKLFAY 503
>TC18700 similar to UP|Q9LYR6 (Q9LYR6) Peptide transporter-like protein,
partial (18%)
Length = 615
Score = 99.4 bits (246), Expect = 1e-21
Identities = 46/118 (38%), Positives = 75/118 (62%)
Frame = +1
Query: 395 ISLFWLSFQYAIFGIADMFTLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVN 454
+S +WL QY + G+A++F +VGLLEF Y EAP MKS+ +++ L+ LG F +T+ +
Sbjct: 46 LSAYWLLIQYCLIGVAEVFCIVGLLEFLYEEAPDAMKSIGSAYAALAGGLGCFAATIINS 225
Query: 455 VINTVTKRITPSKQGWLQGFDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYKSEE 512
+I ++T + K+ WL ++N + FYW L LS +NF F+Y A YKY++++
Sbjct: 226 IIKSLTGK--EGKESWL-AQNINTGRFDYFYWILTALSLVNFCIFIYSAHRYKYRTQQ 390
>AV416871
Length = 354
Score = 98.6 bits (244), Expect = 2e-21
Identities = 55/115 (47%), Positives = 73/115 (62%), Gaps = 11/115 (9%)
Frame = +3
Query: 228 YEVYKDAT----IEKIVHTNQMRFLDKAAILQE------NSESQQPWKVCTVTQVEEVKI 277
YE+ +D IE + H + RFLDKAAI +E ES WK+C VTQVE KI
Sbjct: 3 YEIEQDKEAVMEIEFLPHRDIFRFLDKAAIQRECDMELEKPESTSQWKLCRVTQVENAKI 182
Query: 278 LTRMLPILASTIVMNTCLAQLQTFSVQQGNSMNLKL-GSFTVPASSIPVIPLIFL 331
+ M P+ TI+M CLAQ+QTFSVQQG +M+ K+ F +P +S+P+IP+IFL
Sbjct: 183 ILSMGPVFLCTIIMTLCLAQIQTFSVQQGYTMDTKITKDFNMPPASLPIIPVIFL 347
>TC19180 similar to PIR|T10255|T10255 nitrite transport protein, chloroplast
- cucumber {Cucumis sativus;} , partial (12%)
Length = 408
Score = 96.3 bits (238), Expect = 1e-20
Identities = 50/132 (37%), Positives = 84/132 (62%), Gaps = 1/132 (0%)
Frame = +1
Query: 255 QENSESQQPWKVCTVTQVEEVKILTRMLPILASTIVMNTCLAQLQTFSVQQGNSMNLKLG 314
++NS++ W++ TV +VEE+K + RM PI AS I++ T AQ TFS+QQ +M+ +
Sbjct: 13 EDNSKTPNKWRLNTVHRVEELKSIIRMGPIWASGILLITAYAQQGTFSLQQAKTMDRHIT 192
Query: 315 -SFTVPASSIPVIPLIFLCTLIPIYELFFVPFIRKITHHPSGVTQLQRVGVGLVLSAISM 373
SF +PA S+ V +I + T +Y+ + R+ T G++ L R+G+G V+S I+
Sbjct: 193 KSFQIPAGSMSVFTIITMLTTTALYDRVLIRVARRFTGLDRGISFLHRMGIGFVISTIAT 372
Query: 374 TIAGFIEVKRRD 385
+AGF+E+KR++
Sbjct: 373 FVAGFVEMKRKN 408
>AV409553
Length = 433
Score = 94.4 bits (233), Expect = 5e-20
Identities = 48/135 (35%), Positives = 78/135 (57%), Gaps = 4/135 (2%)
Frame = +2
Query: 65 VMALVMITVQAALDNLHPKA----CGKSSCVEGGIAVMFYTSLCLYALGMGGVRGSLTAF 120
VM ++++TV +L +L P C K+S + V FYT+L A+G GG + +++ F
Sbjct: 29 VMGMILLTVAVSLKSLKPTCTNGICNKASTSQ---IVFFYTALYTTAIGAGGTKPNISTF 199
Query: 121 GANQFDEKDPNEAKALATFFNWLLLSSTLGSVIGVTGIVWVSTQKAWHWGFFIITVASSI 180
GA+QFD+ +P+E + A+FFNW + +S LG++I G+V++ W G+ I T +
Sbjct: 200 GADQFDDFNPHEKQTKASFFNWWMFTSFLGALIATLGLVYIQENLGWGLGYGIPTSGLLL 379
Query: 181 GFVTLAIGKPFYRIK 195
V +G P YR K
Sbjct: 380 SLVIFYVGTPMYRHK 424
>AV768501
Length = 442
Score = 82.8 bits (203), Expect = 1e-16
Identities = 42/105 (40%), Positives = 64/105 (60%)
Frame = -1
Query: 416 VGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSKQGWLQGFD 475
+G L+FF RE P MK++S ++SLG+F S++ V +++ ++T +QGWL +
Sbjct: 442 IGQLDFFLRECPKGMKTMSPGLFLSTLSLGFFFSSMLVTLVH----KMTGPRQGWLAD-N 278
Query: 476 LNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYKSEEDSNNSKEL 520
LNQ L+ FYW LA+LS +N FL+ A WY YK + + EL
Sbjct: 277 LNQGKLDDFYWLLAVLSGVNLVVFLFCAKWYVYKDKRLAQEGIEL 143
>TC8505 similar to UP|Q8LG02 (Q8LG02) Nitrate transporter, partial (23%)
Length = 1495
Score = 81.6 bits (200), Expect = 3e-16
Identities = 52/161 (32%), Positives = 87/161 (53%), Gaps = 3/161 (1%)
Frame = +3
Query: 356 VTQLQRVGVGLVLSAISMTIA-GFIEVKRRDQGRKDPSKPIS--LFWLSFQYAIFGIADM 412
+T L LVL S+ + F+++ R+ G+ ++ LF + +
Sbjct: 69 ITYLTLHSSPLVLDLFSLLLP*WFLQLLRKKGGKMLLRNTLT*VLFGWFLSFFWWEQGKA 248
Query: 413 FTLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITPSKQGWLQ 472
F VG LEFF REAP MKS+ST ++S+GYF+S++ V++++ + SK+ WL+
Sbjct: 249 FAYVGQLEFFIREAPERMKSMSTGLFLAALSMGYFVSSLLVSIVDKL------SKKKWLK 410
Query: 473 GFDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYKSEED 513
+LN+ L+ FYW LA+L LNF F+ A + Y+ + +
Sbjct: 411 S-NLNKGRLDYFYWLLAVLGVLNFILFIVLAMRHHYQVQHN 530
>CB828533
Length = 557
Score = 77.0 bits (188), Expect = 8e-15
Identities = 42/123 (34%), Positives = 62/123 (50%), Gaps = 2/123 (1%)
Frame = +1
Query: 90 CVE--GGIAVMFYTSLCLYALGMGGVRGSLTAFGANQFDEKDPNEAKALATFFNWLLLSS 147
C+E G + +L A+G GG++ S++ FG++QFD DP E K + FFN
Sbjct: 187 CIEASGKQVALLLVALYTIAVGAGGIKSSVSGFGSDQFDTTDPREEKKMVFFFNRFYFFV 366
Query: 148 TLGSVIGVTGIVWVSTQKAWHWGFFIITVASSIGFVTLAIGKPFYRIKTPGESPILRIIQ 207
+ GS+ V +V+V WGF I + L GKP YR K P SP+ I +
Sbjct: 367 STGSLFSVLVLVYVQDNIGRGWGFGIPAGIMLVSLAFLLYGKPLYRYKRPQGSPLTLIWK 546
Query: 208 VIV 210
V++
Sbjct: 547 VLI 555
>BP076619
Length = 432
Score = 73.6 bits (179), Expect = 8e-14
Identities = 38/106 (35%), Positives = 63/106 (58%)
Frame = -3
Query: 406 IFGIADMFTLVGLLEFFYREAPSTMKSLSTSFTFLSMSLGYFLSTVFVNVINTVTKRITP 465
+FG++++FTLVGL EFFY + P+ ++SL + +G FLS ++VI TVT + P
Sbjct: 427 LFGVSEVFTLVGLQEFFYDQVPNELRSLGLALYLSIFGVGSFLSGFLISVIETVTGKDGP 248
Query: 466 SKQGWLQGFDLNQNNLNLFYWFLAILSCLNFFNFLYWASWYKYKSE 511
+ W +L++ +++ FYW LA S + F F+ +A Y Y +
Sbjct: 247 DR--WFAN-NLHKAHIDYFYWLLAGFSVVGFAMFMCFAKSYVYNHQ 119
>BI420182
Length = 506
Score = 70.9 bits (172), Expect = 5e-13
Identities = 41/102 (40%), Positives = 59/102 (57%), Gaps = 4/102 (3%)
Frame = +3
Query: 1 MGFVTNMVSLVLYFIGVMHFDLSSSANTLTNFMGSTFLLSLVGAFISDTYLNRFTTCLVF 60
+GF TNM+S Y +H L+ +ANTLTNF G+ L L+GAFISD+Y +F T +
Sbjct: 210 VGFSTNMIS---YLTTQLHMPLTKAANTLTNFGGTASLTPLLGAFISDSYAGKFWTITMA 380
Query: 61 GSLEVMALVMITVQAALDNLHPKAC-GKSSC---VEGGIAVM 98
L + +V +T+ A L L P C G+ C +G +AV+
Sbjct: 381 SVLYQIGMVSLTISAVLPQLRPPPCRGEEVCKQATDGQLAVL 506
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.323 0.137 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,126,891
Number of Sequences: 28460
Number of extensions: 146289
Number of successful extensions: 1042
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 1001
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1005
length of query: 569
length of database: 4,897,600
effective HSP length: 95
effective length of query: 474
effective length of database: 2,193,900
effective search space: 1039908600
effective search space used: 1039908600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148406.6


