

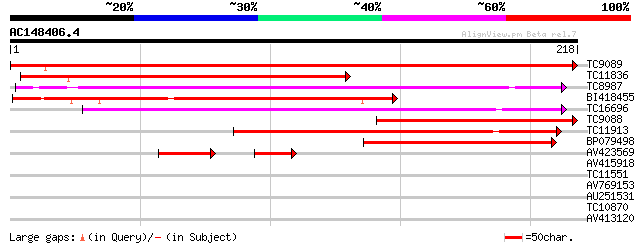
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148406.4 - phase: 0
(218 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC9089 similar to UP|RHRE_PEA (Q9S8P4) Rhicadhesin receptor prec... 365 e-102
TC11836 similar to GB|AAB51577.1|1755176|ATU75199 germin-like pr... 195 4e-51
TC8987 similar to UP|ABPA_PRUPE (Q9ZRA4) Auxin-binding protein A... 147 1e-36
BI418455 144 2e-35
TC16696 similar to UP|Q9AR81 (Q9AR81) Germin-like protein precur... 129 3e-31
TC9088 similar to UP|RHRE_PEA (Q9S8P4) Rhicadhesin receptor prec... 120 2e-28
TC11913 weakly similar to UP|ABPA_PRUPE (Q9ZRA4) Auxin-binding p... 95 8e-21
BP079498 62 6e-11
AV423569 41 1e-06
AV415918 32 0.11
TC11551 homologue to UP|AAQ93070 (AAQ93070) 3-ketoacyl-CoA thiol... 30 0.32
AV769153 28 0.93
AU251531 26 4.6
TC10870 similar to GB|CAA77438.1|2924285|CHNTXX Ycf2 protein {Ni... 26 6.1
AV413120 25 7.9
>TC9089 similar to UP|RHRE_PEA (Q9S8P4) Rhicadhesin receptor precursor
(Germin-like protein), partial (93%)
Length = 981
Score = 365 bits (936), Expect = e-102
Identities = 188/220 (85%), Positives = 200/220 (90%), Gaps = 2/220 (0%)
Frame = +1
Query: 1 MKLEHSLFLGTL--ALLLATSFSSDPDYLQDLCVADLASGVTVNGFTCKEASKVNAFDFS 58
MKL SL L L ALLL T+FSSDPDYLQDLCVAD ASGV VNGFTCKEASKVNA DF
Sbjct: 154 MKLVDSLLLFALPLALLLTTAFSSDPDYLQDLCVADPASGVPVNGFTCKEASKVNASDFF 333
Query: 59 SIILAKPGSTNNTFGSVVTGANVQKVPGLNTLGVSLSRIDYAPGGLNPPHTHPRATEVVF 118
S ILAKPG+TNNTFGS+VTGANVQKVPGLNTLGVSL+RIDYAP GLNPPHTHPRATEVVF
Sbjct: 334 SNILAKPGATNNTFGSLVTGANVQKVPGLNTLGVSLARIDYAPDGLNPPHTHPRATEVVF 513
Query: 119 VLEGQLDVGFITTANVLISKTISKGEIFVFPKGLVHFQKNNANVPASVLSAFNSQLPGTQ 178
VLEG+LDVGFITT NVLISK I KG+IFVFPKGL+HFQKN VPA+VLSAFNSQLPGTQ
Sbjct: 514 VLEGELDVGFITTGNVLISKHIVKGDIFVFPKGLLHFQKNXGKVPAAVLSAFNSQLPGTQ 693
Query: 179 SIATTLFAATPSVPDNVLTKTFQVGTKEVEKIKSRLAPKK 218
SIATTLFA+TP+VPDNVLTKTFQVGTK++EKIKSRLAPKK
Sbjct: 694 SIATTLFASTPTVPDNVLTKTFQVGTKQIEKIKSRLAPKK 813
>TC11836 similar to GB|AAB51577.1|1755176|ATU75199 germin-like protein
{Arabidopsis thaliana;} , partial (56%)
Length = 434
Score = 195 bits (496), Expect = 4e-51
Identities = 98/129 (75%), Positives = 108/129 (82%), Gaps = 2/129 (1%)
Frame = +3
Query: 5 HSLFLGTLALLLATSFS--SDPDYLQDLCVADLASGVTVNGFTCKEASKVNAFDFSSIIL 62
H + L L LL T+F+ SDPD LQD+CVADL+S + VNGF CK AS V DF S IL
Sbjct: 48 HIMKLVVLLLLTLTTFTAASDPDSLQDICVADLSSAIKVNGFVCKSASAVKETDFFSAIL 227
Query: 63 AKPGSTNNTFGSVVTGANVQKVPGLNTLGVSLSRIDYAPGGLNPPHTHPRATEVVFVLEG 122
AKPG+TN TFGSVVTGANV+K+PGLNTLGVS+SRIDYAPGGLNPPHTHPRATEVVFVLEG
Sbjct: 228 AKPGATNTTFGSVVTGANVEKIPGLNTLGVSISRIDYAPGGLNPPHTHPRATEVVFVLEG 407
Query: 123 QLDVGFITT 131
QLDVGFITT
Sbjct: 408 QLDVGFITT 434
>TC8987 similar to UP|ABPA_PRUPE (Q9ZRA4) Auxin-binding protein ABP19a
precursor, partial (97%)
Length = 985
Score = 147 bits (371), Expect = 1e-36
Identities = 86/212 (40%), Positives = 125/212 (58%)
Frame = +1
Query: 3 LEHSLFLGTLALLLATSFSSDPDYLQDLCVADLASGVTVNGFTCKEASKVNAFDFSSIIL 62
L H +FL +LLL+TS ++ +QD CVADL G+ CK + V + DF L
Sbjct: 28 LPHIVFL--FSLLLSTSHAA----VQDFCVADLKGSDGPAGYPCKTPATVTSDDFVFAGL 189
Query: 63 AKPGSTNNTFGSVVTGANVQKVPGLNTLGVSLSRIDYAPGGLNPPHTHPRATEVVFVLEG 122
+ PG+ N G+ VT A V + PG+N LG+S +R+D PGG+ P HTHP A E++ V +G
Sbjct: 190 SAPGNITNIIGAAVTPAFVGQFPGVNGLGLSAARLDIGPGGVIPLHTHPGANELLIVTQG 369
Query: 123 QLDVGFITTANVLISKTISKGEIFVFPKGLVHFQKNNANVPASVLSAFNSQLPGTQSIAT 182
+ GFI++A+ + KT+ KGE+ VFP+GL+HFQ A + F+S PG Q +
Sbjct: 370 HITAGFISSASTVYIKTLKKGELMVFPQGLLHFQVAAGKTKAIGFAIFSSASPGLQILDF 549
Query: 183 TLFAATPSVPDNVLTKTFQVGTKEVEKIKSRL 214
LFA+ S P ++TKT + V+K+K L
Sbjct: 550 ALFASNFSTP--LITKTTFLDPALVKKLKGVL 639
>BI418455
Length = 477
Score = 144 bits (362), Expect = 2e-35
Identities = 83/157 (52%), Positives = 108/157 (67%), Gaps = 9/157 (5%)
Frame = +1
Query: 2 KLEHSLFLGTLALLLATSFSS--DPDYLQDLCVA--DLASGVTVNGFTCKEASKVNAFDF 57
K+E FL T+ L LA+SF+S DP LQD CV D GV VNG CK+ + V A DF
Sbjct: 13 KMEVVYFLVTI-LALASSFASAYDPSPLQDFCVGINDPKYGVFVNGKFCKDPALVKAEDF 189
Query: 58 SSIILAKPGSTNNTFGSVVTGANVQKVPGLNTLGVSLSRIDYAPGGLNPPHTHPRATEVV 117
+ +PG+T+N GS VT V+++ GLNTLG+SL+RID+AP GLNPPHTHPRATE++
Sbjct: 190 FKHV--EPGNTSNPLGSKVTPVTVEQLFGLNTLGISLARIDFAPKGLNPPHTHPRATEIL 363
Query: 118 FVLEGQLDVGFITTANV-----LISKTISKGEIFVFP 149
VLEG L VGF+T+ N L +K ++KG++FVFP
Sbjct: 364 IVLEGTLYVGFVTSNNANGGNRLFTKVLNKGDVFVFP 474
>TC16696 similar to UP|Q9AR81 (Q9AR81) Germin-like protein precursor,
partial (96%)
Length = 834
Score = 129 bits (325), Expect = 3e-31
Identities = 70/186 (37%), Positives = 108/186 (57%)
Frame = +1
Query: 29 DLCVADLASGVTVNGFTCKEASKVNAFDFSSIILAKPGSTNNTFGSVVTGANVQKVPGLN 88
D CV DL G+ CK+ SKV A DF+ L G+T+ + VT A + PG+N
Sbjct: 55 DFCVGDLTFPNGPAGYACKKPSKVTADDFAYSGLGIAGNTSYIIKAAVTPAFDAQFPGVN 234
Query: 89 TLGVSLSRIDYAPGGLNPPHTHPRATEVVFVLEGQLDVGFITTANVLISKTISKGEIFVF 148
LG+SL+R+D A GG+ P HTHP A+E + V++G + GF+ + N + KT+ KG++ VF
Sbjct: 235 GLGISLARLDLASGGVIPLHTHPGASEALVVVQGTICAGFVASDNTVYLKTLKKGDVMVF 414
Query: 149 PKGLVHFQKNNANVPASVLSAFNSQLPGTQSIATTLFAATPSVPDNVLTKTFQVGTKEVE 208
P+GLVHFQ ++ A +F+S PG Q + LF + P +++ T + +++
Sbjct: 415 PQGLVHFQIHDGGSSALAFVSFSSANPGLQILDFALFKS--DFPSDLIAATAFLDAAQIK 588
Query: 209 KIKSRL 214
K+K L
Sbjct: 589 KLKGVL 606
>TC9088 similar to UP|RHRE_PEA (Q9S8P4) Rhicadhesin receptor precursor
(Germin-like protein), partial (35%)
Length = 475
Score = 120 bits (301), Expect = 2e-28
Identities = 60/77 (77%), Positives = 64/77 (82%)
Frame = +3
Query: 142 KGEIFVFPKGLVHFQKNNANVPASVLSAFNSQLPGTQSIATTLFAATPSVPDNVLTKTFQ 201
KGEIFVFPKGLVH+QKNN PASV+SAFNSQLPGT SI LFAA P VPDNVLTK FQ
Sbjct: 3 KGEIFVFPKGLVHYQKNNGYWPASVISAFNSQLPGTVSIPLALFAAKPPVPDNVLTKAFQ 182
Query: 202 VGTKEVEKIKSRLAPKK 218
T+EVEKIK+ LAPKK
Sbjct: 183 TSTEEVEKIKANLAPKK 233
>TC11913 weakly similar to UP|ABPA_PRUPE (Q9ZRA4) Auxin-binding protein
ABP19a precursor, partial (61%)
Length = 530
Score = 95.1 bits (235), Expect = 8e-21
Identities = 48/126 (38%), Positives = 79/126 (62%)
Frame = +1
Query: 87 LNTLGVSLSRIDYAPGGLNPPHTHPRATEVVFVLEGQLDVGFITTANVLISKTISKGEIF 146
+N LG+S +R+D GG+ P H+HP A+E+V +L+G++ VGFI+T N + KT+ KG+I
Sbjct: 16 INGLGLSAARLDLDAGGVVPIHSHPGASELVIILQGRITVGFISTDNTVYQKTLMKGDII 195
Query: 147 VFPKGLVHFQKNNANVPASVLSAFNSQLPGTQSIATTLFAATPSVPDNVLTKTFQVGTKE 206
V P+GL+HFQ N AS + F+S P Q + LF ++ ++ +T + +
Sbjct: 196 VIPQGLLHFQLNAGGNRASAVLTFSSTNPVAQLVDVALFG--NNLDSGLVARTTFLDLAQ 369
Query: 207 VEKIKS 212
V+K+K+
Sbjct: 370 VKKLKA 387
>BP079498
Length = 373
Score = 62.4 bits (150), Expect = 6e-11
Identities = 29/74 (39%), Positives = 45/74 (60%)
Frame = -2
Query: 137 SKTISKGEIFVFPKGLVHFQKNNANVPASVLSAFNSQLPGTQSIATTLFAATPSVPDNVL 196
+K ++KG++FVFP L+HFQ N + A ++ +SQ PG +IA LF A+P + VL
Sbjct: 372 TKVLNKGDVFVFPISLIHFQLNVGSGNAVAIAGLSSQNPGVITIANALFKASPPISQEVL 193
Query: 197 TKTFQVGTKEVEKI 210
TK QV ++ +
Sbjct: 192 TKALQVDKSIIDNL 151
>AV423569
Length = 351
Score = 40.8 bits (94), Expect(2) = 1e-06
Identities = 16/16 (100%), Positives = 16/16 (100%)
Frame = +3
Query: 95 SRIDYAPGGLNPPHTH 110
SRIDYAPGGLNPPHTH
Sbjct: 303 SRIDYAPGGLNPPHTH 350
Score = 26.9 bits (58), Expect(2) = 1e-06
Identities = 13/22 (59%), Positives = 16/22 (72%)
Frame = +2
Query: 58 SSIILAKPGSTNNTFGSVVTGA 79
SS ++ +TN TFGSVVTGA
Sbjct: 200 SSHHTSQTSATNTTFGSVVTGA 265
>AV415918
Length = 189
Score = 31.6 bits (70), Expect = 0.11
Identities = 24/66 (36%), Positives = 34/66 (51%)
Frame = +3
Query: 7 LFLGTLALLLATSFSSDPDYLQDLCVADLASGVTVNGFTCKEASKVNAFDFSSIILAKPG 66
LFL T L TS++S + DLCVAD T +G+ C + + + DF L G
Sbjct: 9 LFLFTF--LSTTSYAS----VNDLCVADFKGPSTPSGYHCMPLANLTSDDFVFHGLV-AG 167
Query: 67 STNNTF 72
+T N+F
Sbjct: 168NTTNSF 185
>TC11551 homologue to UP|AAQ93070 (AAQ93070) 3-ketoacyl-CoA thiolase ,
partial (37%)
Length = 915
Score = 30.0 bits (66), Expect = 0.32
Identities = 21/75 (28%), Positives = 34/75 (45%)
Frame = -2
Query: 15 LLATSFSSDPDYLQDLCVADLASGVTVNGFTCKEASKVNAFDFSSIILAKPGSTNNTFGS 74
L T S P++LQ+ D + + N + DF++ +AK G T T GS
Sbjct: 329 LTLTFLVSSPNFLQE*TNCDANASLISNKSMLPSSRPA---DFTAAGIAKAGPTPMTEGS 159
Query: 75 VVTGANVQKVPGLNT 89
T A + K+P + +
Sbjct: 158 TPTAAKLLKIPRIGS 114
>AV769153
Length = 435
Score = 28.5 bits (62), Expect = 0.93
Identities = 14/46 (30%), Positives = 26/46 (56%)
Frame = -1
Query: 7 LFLGTLALLLATSFSSDPDYLQDLCVADLASGVTVNGFTCKEASKV 52
LF L+L+ +TS +++ CV+ L SG+T +G +E + +
Sbjct: 261 LFTAVLSLINSTS-EGLANHISQFCVSHLTSGITTDGEVLRETTLI 127
>AU251531
Length = 400
Score = 26.2 bits (56), Expect = 4.6
Identities = 15/40 (37%), Positives = 21/40 (52%), Gaps = 2/40 (5%)
Frame = +3
Query: 163 PASVLSAFNS--QLPGTQSIATTLFAATPSVPDNVLTKTF 200
P S L A + QLP S + TL +TPS+P + T+
Sbjct: 48 PFSSLPALSCILQLPTHNSFSLTLLVSTPSIPCAICKATY 167
>TC10870 similar to GB|CAA77438.1|2924285|CHNTXX Ycf2 protein {Nicotiana
tabacum;}, complete
Length = 6897
Score = 25.8 bits (55), Expect = 6.1
Identities = 12/28 (42%), Positives = 17/28 (59%)
Frame = +1
Query: 70 NTFGSVVTGANVQKVPGLNTLGVSLSRI 97
N FGS+ G+NVQ + L +S+S I
Sbjct: 5530 NGFGSITLGSNVQDLVALTNEALSISII 5613
>AV413120
Length = 330
Score = 25.4 bits (54), Expect = 7.9
Identities = 19/49 (38%), Positives = 25/49 (50%), Gaps = 3/49 (6%)
Frame = -2
Query: 5 HSLFLGTLALLLATSFSSDPDYLQDLCVADLASGVTVN---GFTCKEAS 50
H L L L LLL + SSD D L VA +A+ V + G+ C E +
Sbjct: 239 HGLCLTPLYLLLLSPLSSDGDADASLDVAVVAASVAEDDDCGWDCCEGT 93
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.133 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,140,634
Number of Sequences: 28460
Number of extensions: 37287
Number of successful extensions: 194
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 191
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 191
length of query: 218
length of database: 4,897,600
effective HSP length: 87
effective length of query: 131
effective length of database: 2,421,580
effective search space: 317226980
effective search space used: 317226980
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 53 (25.0 bits)
Medicago: description of AC148406.4


